Translational Research on Amyotrophic Lateral Sclerosis (ALS): The Preclinical SOD1 Mouse Model
Maria Mina, Eleni Konsolaki and Laskaro Zagoraiou*
DOI10.21767/2573-5349.100022
Maria Mina1,2, Eleni Konsolaki2 and Laskaro Zagoraiou2*
1Department of Basic Sciences, Faculty of Nursing, University of Athens, 123 Papadiamantopoulou Str., 11527, Athens, Greece
2Center of Basic Research, Biomedical Research Foundation of the Academy of Athens, 4 Soranou Ephessiou Str., 11527, Athens, Greece
- *Corresponding Author:
- Laskaro Zagoraiou
Center of Basic Research, Biomedical Research Foundation of the Academy of Athens
4 Soranou Ephessiou Str., 11527, Athens, Greece
Tel: 00302106597514
Fax: 00302106597545
E-mail: lzagoraiou@bioacademy.gr
Received Date: July 20, 2018; Accepted Date: August 07, 2018; Published Date: August 14, 2018
Citation: Mina M, Konsolaki E, Zagoraiou L (2018) Translational Research on Amyotrophic Lateral Sclerosis (ALS): The Preclinical SOD1 Mouse Model. J Transl Neurosci 3:9. DOI: 10.21767/2573-5349.100022
Abstract
Amyotrophic Lateral Sclerosis (ALS) is the most common motor neuron disease and is clinically defined by the degeneration of upper and lower motor neurons, leading to paralysis and premature death. 10% of ALS patients suffer from the familial form of the disease and research has revealed a number of responsible mutations in specific genes and loci. Transgenic mouse models carrying human ALS related mutations have been generated for the study of the mechanisms involved in the disease pathogenesis and putative therapies. Mutations in the Cu/ Zn superoxide dismutase 1 (SOD1) gene, accounting for 20% of fALS and up to 5% of sALS, were the first identified to be involved in the disease. In this review, we focus on the first transgenic model used, the SOD1 mouse, which recapitulates many symptoms of the human ALS pathology. SOD1 mouse models have been extensively studied in basic and translational research, in order to unravel the underlying mechanisms, the early signs of the disorder and potential therapeutic interventions. In basic research this model has provided valuable information about ALS causes and progression. In translational research, encouraging results have emerged, but the need for better design of clinical trials is evident. This review presents the impact of the SOD1 mouse models in ALS investigation.
Keywords
ALS; SOD1; ALS mouse models; Translational research; Drugs
Introduction
Amyotrophic Lateral Sclerosis (ALS), also known as Lou Gehrig's disease, is a heterogeneous fatal neurodegenerative disease that belongs to a wider group of disorders known as motor neuron diseases [1]. It is clinically defined by the degeneration of both upper and lower motor neurons leading, to paralysis and death usually within 2 to 5 years after its onset [2] , although some individuals show protracted survival. The 90% of ALS patients suffer from the sporadic form of the disease (sALS), while the 10% suffer from familial ALS (fALS). The incidence of ALS is approximately 1-2.6 per 100 000 persons per year and its prevalence is 6 cases per 100.000 [3]. Although quite rare in the population, ALS is the most common motor neuron disease in adults, so research on the underlying mechanisms and potential therapeutic interventions would be of immense clinical importance.
Clinical Phenotypes and Genetics in ALS Patients
Typically, the onset of the disease occurs at a mean age of 55 years, starting focally with different sets of motor neurons and different regions of the body affected. Sporadic and familial ALS present similar pathological and clinical profiles [4]. The current clinical standard for diagnosis of ALS is based on the Revised “El Escorial World Federation of Neurology Criteria”; however, no single test confirms the diagnosis of ALS [2]. A combination of clinical examination, neuroimaging and electrophysiological tests aids the diagnosis. Three classes of patients could be detected based on the onset of the disease and the type of motor neuron affected [5,6]: i) Spinal onset (70% of the patients): involves weakness in the limbs attributed to degeneration of lower motor neurons (neurons from the brainstem or spinal cord that project to muscle) or hyperreflexia due to dysfunction of upper motor neurons (neurons from the cortex which project to the brainstem and the spinal cord); ii) Bulbar onset (25% of patients): includes tongue atrophy with thickness of speech and difficulty in swallowing due to brainstem motor neuron degeneration; iii) Respiratory onset (5% of the patients): the least common pattern in which the brainstem neurons and respiratory system are affected. The respiratory muscle dysfunction represents the terminal phase of the disease in all classes.
Research on postmortem tissues from ALS patients has revealed many neuropathological events that lead to the degeneration of upper and lower motor neurons and eventually to the disease symptoms. These events take place in the motor neurons and include cytoplasmic protein aggregates, glutamate-induced excitotoxicity, mitochondrial abnormalities, dysfunction of axonal transport, altered RNA processing, proteasomal dysfunction and generation of free radicals [5]. Furthermore, astrocyte and microglia activation has been reported [7] and may also contribute to the loss of motor neurons by releasing toxic factors and insufficiently removing glutamate from the synaptic cleft, causing excitotoxicity [8]. Combination of some or all the above events results in activation of calcium-dependent pathways and increased oxidative stress inside the motor neuron, leading to its final degeneration.
Many different mechanisms have been implicated to the disease generation and no cure has been found yet, since the anti-glutamatergic agent riluzole and the anti-oxidant edaravone may only extend lifespan up to some months. Thus, research on the genes accounting for ALS and possible environmental risk factors is of high importance in order to understand the causes of the disease. Environmental risk factors include: physical activity, smoking and exposure to heavy metals, pesticides and chemicals [9]. Over the last 25 years, research οn ALS patients revealed a number of genes and loci related to the disease. Mutations in the Cu/Zn superoxide dismutase 1 (SOD1) gene were the first identified to be involved in the disease [10], accounting for 20% of fALS and up to 5% of sALS. SOD1 is an enzyme that breaks down superoxide radicals, which are toxic for many cellular components. Up to date, more than 160 missense mutations of SOD1 gene have been found to be linked to ALS. The next gene discovered in patients was the TARDBP/TDP-43 gene encoding for TAR DNA-binding protein (TARDBP or TDP-43) [11,12], mutations in which account for almost 5% of fALS cases. TDP-43 protein is normally found in the nucleus and plays a role in RNA processing and transport [13]. At the same time, mutations of the gene encoding for angiogenin (ANG), an RNase A involved in angiogenesis, were found in some patients of fALS and sALS [14]. Soon after the discovery of TARDBP/TDP-43 mutations, the gene encoding for fused in sarcoma (FUS), another RNA processing protein, was also identified as a gene whose mutations result in almost 4% of fALS and rare sALS cases [15,16,17]. Subsequently, mutations of OPTN gene coding for optineurin, a protein involved in autophagy, were also linked to some ALS cases [18]. In 2011, the involvement of an expanded nucleotide repeat in ALS was revealed for the first time. More specifically, expansion of a non-coding hexanucleotide repeat (GGGGCC) in the C9ORF72 gene was identified in 35-45% of fALS and 10% of sALS patients [19,20], leading to reduced levels of one alternatively spliced C9ORF72 transcript. The presence of transcripts accumulation as nuclear RNA foci though, favors a toxic RNA gain of function mechanism. Healthy individuals carry up to 20 repeats, while the presence of greater than 30 repeats is considered pathogenic [20]. Mutations in several other genes have also been identified in ALS cases, but they are quite rare and their involvement in the disease needs further investigation. Two examples are the mutations of ubiquilin 2 (UBQLN2) [21] and sequestosome 1 (SQSTM1 or p62) [22] genes, both involved in the degradation of proteins by the proteasome and in autophagy process.
Transgenic Mouse Models in ALS Research
Several transgenic mouse models, harboring some of the known mutations or the hexanucleotide repeat expansion in ALS patients, have been generated for the study of the mechanisms involved in the disease pathogenesis. The first model used was the SOD1 mouse line carrying the human G93A mutation [23], which was found to recapitulate many of the ALS symptoms. Today there are more than 12 different SOD1 mouse mutants that have several human missense mutations and result in many ALS hallmarks, such as development of SOD1 cytoplasmic aggregates and severe motor deficits.
The next transgenic mouse line generated was the TDP43 model, which has been used over 20 years to provide an opportunity to study RNA processing as a potential disease mechanism, leading to motor neuron degeneration. Many research groups have used mouse models overexpressing either the human wild type TDP43 (hTDP-43WT) or one of its ALS-correlated mutated forms that have been recognized in patients (M337V, A315T, Q331K, G348C), under the control of mainly three different promoters. More specifically, some transgenic mice ubiquitously overexpress hTDP-43WT [24-27] or hTDP43 carrying the mutations M337V [24,26,28] , A315T [24,29] , Q331K [26,27] under the control of murine prion promoter (mPrP), which results in higher expression in the CNS and heart, and lower at skeletal muscle, liver and kidney [30]. In other mouse lines, human wild type [31,32] or mutated TDP-43 (M337V) are overexpressed in neurons, under the control of murine Thy1.2 promoter [33,34]. The last promoter, under which the hTDP-43WT [35] is overexpressed in the neurons of the hippocampus and cortex of transgenic mice, is the murine CaMK2 [36]. These transgenic TDP43 mouse models present various pathogenic characteristics depending on the type of promoter and levels of expression, such as dose dependent toxicity of TDP43 protein, cytoplasmic TDP43 aggregates, progressive paralysis and death, but none of these mice recapitulate the ALS phenotype adequately. Except from the investigation of the effects of TDP43 overexpression, some groups studied also the possible ALS-related phenotypes due to loss of function of this protein. The homozygous Tdp43 knockout mice usually die around embryonic day 7, pointing to the importance of TDP43 protein for normal development [37- 39]. The heterozygous Tdp43 knockout mice do survive and do not demonstrate a reduced TDP43 expression, but they develop age-related motor and muscle dysfunction [37]. Generation of conditional Tdp43 knockout mice showed immediate excessive weight loss and death after the postnatal reduction of TDP43 expression (9 days after the application of tamoxifen), due to increased fat oxidation [40]. Taken together, none of the TDP43 transgenic models appear to develop enough ALS hallmarks, indicating that abnormal TDP43 in mice may be not sufficient for the recapitulation of the disease.
Genetically engineered Fused in sarcoma (FUS) mice were also generated to investigate the loss and gain of function mechanisms leading in ALS. The first Fus knock out mice generated died perinataly (24 hours after birth) [41]. Outbred Fus knock out mice survive and show behavioral and pathological abnormalities, which do not correlate to ALS [42]. However, overexpression of human wild type FUS was found to lead to degeneration of motor neurons and paralysis [43]. Quite recently, Sharma et al used transgenic mice which conditionally express the human wild type or the ALS-associated mutant (R521C or P525L) FUS and showed that mutant FUS is the one responsible for the degeneration of motor neurons through its toxic gain of function, without the involvement of normal FUS [44].
Since the expansion of a specific repeat (GGGGCC) in the C9ORF72 gene was found to account for the largest percentage of fALS cases and the patients present also elevated antisense transcripts, mice having increased number of this hexanucleotide repeat and expressing sense and antisense transcripts were established. Chew et al. injected the adeno-associated virus (AAV), expressing 66 repeats, into the CNS of mice [45]. These mice exhibit some of the ALS features such as nuclear RNA foci, phospho-TDP43 inclusions, dipeptide protein repeat inclusions, motor impairment and cognitive symptoms, like anxiety and social abnormalities. Other mouse models were generated to harbor a bacterial artificial chromosome (BAC) carrying 500 [46] or 100-1000 [47] repeats. These mice only showed a few pathogenic abnormalities, but not any motor or behavioral phenotype. Recently, two more groups generated transgenic mice, in which BACs containing 450 [48] and 500 [49] repeats were delivered. The mice carrying 450 repeats developed some ALS neuropathological characteristics such as RNA foci, dipeptide protein repeat inclusions, protein accumulation and cognitive deficits, but very mild neurodegeneration. However, the mice carrying 500 repeats developed many of the neuropathological hallmarks of the disease, such as severe motor neuron degeneration and paralysis, as well as cognitive symptoms. The difference in ALS recapitulation between the last model and the previous ones may rely on differences in individual BAC transgenics, in the background of mice and in transgene transcription expression levels. Knocking out the C9orf72 gene from mice did not have any pathological effect on motor neurons, showing no correlation of the loss of function of this gene with ALS [50,51]. Thus, ALS pathology can result from a toxic gain of function acquired from different degree of expansion of the non-coding hexanucleotide repeat in the C9ORF72 gene. This provides a unique causative example, since it is not a missense mutation of a gene, like the other cases described. It opens the way for major progress in basic and translational research in the future.
The rest of the genes whose mutations have been identified in ALS patients, account only for minor percentage of fALS cases and the transgenic mouse models generated for some of them either haven’t been extensively studied yet or haven’t presented any overt motor neuron phenotype. For example, mice carrying the human mutated UBQLN2 gene, although presenting protein aggregates, do not exhibit any motor neuron degeneration [52].
The present review focuses on the ongoing research on mutations of the responsible genes for ALS pathology. The most studied transgenic mouse models carrying mutated causative genes are the SOD1 and TDP43 mice. Since mimicking of the disease symptoms in TDP43 models is limited, we will focus on the SOD1 mice, the first generated animal models for ALS, which carry the most common missense mutations of the fALS cases (20%) and represent human ALS pathology quite well.
Superoxide Dismutase 1 (SOD1) Mouse Models
Mutations of the SOD1 gene are considered as the largest missense mutation yet known to account for the familial (although only 20%) and some sporadic ALS cases in humans [10]. In fact, more than 160 missense mutations have been identified in the human SOD1 gene including G93A, G37R, D90A, A4V, H46R, L84V, and G85R. Since most of the pathophysiological evidence for patients is coming from postmortem tissues and the results correspond mainly to the end stage of the disease, the use of animal models is essential to unravel the responsible mechanisms for the degeneration of motor neurons and paralysis. Due to the conservation of molecular and cellular pathways between humans and mice, transgenic mouse models for ALS have been extensively used, with the SOD1G93A [23] mice being the first ones.
A toxic gain of function of SOD1 seems to have a strong implication to the generation of ALS. Mouse models overexpressing the human wild type and/or the mutated SOD1 gene carrying one of the known ALS-related mutations have been generated and extensively used for the study of the mechanisms responsible for ALS pathogenesis. Transgenic mouse models expressing mutated SOD1 forms in similar or elevated levels as to the endogenous mouse protein are found to recapitulate the ALS-phenotype in a great extent. Up to date, more than 15 different transgenic SOD1 mouse lines have been generated and their main features are summarized in Table 1. Almost all of these mice develop severe degeneration of motor neurons, leading to progressive paralysis of the hindlimbs and forelimbs and death, as seen in ALS human patients. They also present some of the disease main pathophysiological characteristics seen already in postmortem tissues from human patients, such as SOD1 cytoplasmic inclusions, gliosis, glutamate excitotoxicity, vacuolization of mitochondria and disrupted axonal transport. Furthermore, participation of glial cells in the disease pathogenesis was identified for the first time in transgenic mice expressing mutant SOD1 as reviewed by Philips and Rothstein, 2014. More specifically, the surrounding astrocytes and microglia are found to provide less support and to promote glutamate excitotoxicity in the motor neurons by expressing reduced glutamate transporters [53]. Importantly, overexpression of mutant SOD1 only in motor neurons [54,55] or only in astrocytes [56] or Schwann cells [57], failed to result in motor neuron death, indicating ALS as a non-cell autonomous disease. Thus, expression of mutant SOD1 in motor neurons and glial cells at the same time is required for the generation of the disease symptoms. The recapitulation of the disease symptoms and especially the age of onset depends on the type of SOD1 mutation, the level of its expression and the genetic background and sex of the transgenic mouse [58].
| Mutation | Disease onset (months) | Paralysis | Neuropathological findings | Disease duration (months) | References |
|---|---|---|---|---|---|
| G93A | 3-4 | YES | motor neuron degeneration, disrupted neuromuscular junctions, SOD1 inclusions | 1-2 | Gurney et al. [23] |
| G37R | 4-6 | YES | Membrane-bounded vacuoles, motor neuron degeneration, muscle atrophy | 1-3 | Wong et al. [116] |
| G86Ra,b | 3-4 | YES | motor neuron degeneration | 0.5-1 | Ripps et al. [117] |
| G85Rb | 8-14 | YES | SOD1 inclusions in astrocytes and neurons, motor neuron degeneration | 0.5 | Bruijn et al. [53] |
| L84V | 5-6.5 | YES | overexpression of the ER-resident chaperone GRP78/BiP before onset, motor neuron degeneration | 1 | Tobisawa et al. [118] |
| G127Xb | 8 | YES | motor neuron degeneration, SOD1 inclusions | 0.25 | Jonsson et al. [119] |
| H46R | 5 | YES | motor neuron degeneration, SOD1 inclusions, ubiquitin inclusions, Lewy body-like inclusions | 1 | Chang-Hong et al. [120] |
| D90A | 12 | YES | urinary bladder distention, motor neuron degeneration, SOD1 inclusions | 2 | Jonsson et al. [121] |
| L126Z | 7-11 | YES | SOD1 inclusions, motor neuron degeneration | 0.75-3 | Wang et al., Deng et al. [122,73] |
| A4Vc | NO | not defined | Deng et al. [73] | ||
| A4V/SOD1WT | 8.5 | YES | motor neuron degeneration, SOD1 inclusions | 3.5 | Deng et al. [73] |
amouse mutation
bRapid progression
cno ALS-like symptoms
Table 1: Most common mutant SOD1 mouse models.
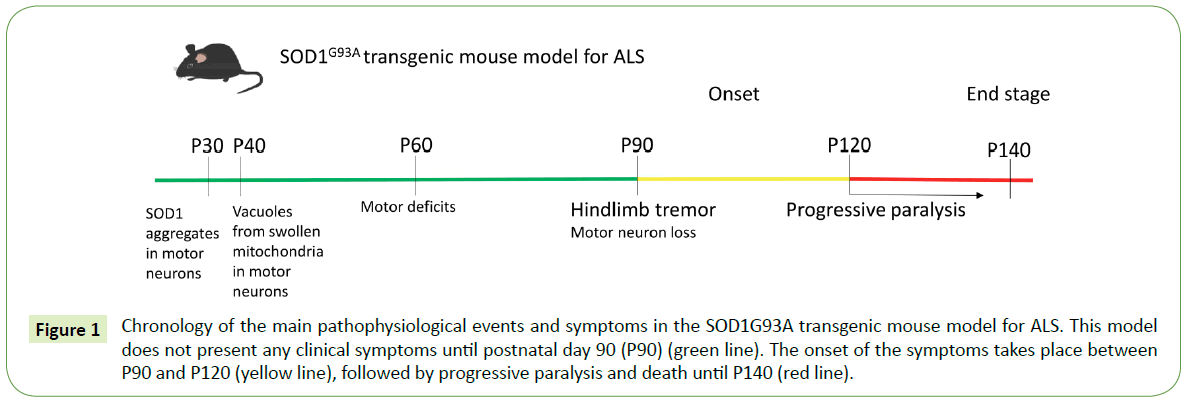
The first transgenic mouse model established was the SOD1G93A overexpressing the human mutation G93A, a substitution of glycine93 to alanine. The SOD1G93A mouse line recapitulates quite many symptoms of the disease and therefore it is the most studied transgenic model for ALS [23] (Figure 1). Several different lines of this mouse model carrying different transgene copy numbers have been established, namely G1, G5, G12 and G20 [23,59,60] , with G1 being the most studied. In these mice, the disease onset takes place at around 3-4 months of age starting with the tremor of hindlimbs and muscle weakness and followed by more intense locomotor deficits, paralysis and death at around 5 months of age [23]. The pathophysiological events at the cellular level that result in the severe loss of spinal motor neurons reported in these mice, are evident before the onset of motor symptoms. One of the first pathological characteristics, the mitochondrial vacuolization and dysfunction [61], is probably caused by SOD1 accumulation inside the mitochondria [62] and is implicated in the ensuing motor neuron degeneration. Enhanced oxidative stress and increased free radicals [63] and glutamate excitotoxicity [64], major causes for motor neuron damage in ALS, are also evident from the early disease stages in SOD1G93A mouse lines. At the same time, axonal transport is found to be seriously disrupted [65] and neuromuscular junctions are impaired and subsequently degenerate [66,67]. Cytosolic SOD1 aggregates present a decreased ubiquitination as the disease progresses [68]. Additionally, activation of glial cells, mainly astrocytes and microglia, after the onset of the symptoms causes gliosis and contributes to an even more intense degeneration of the neighboring motor neurons [69].
Figure 1: Chronology of the main pathophysiological events and symptoms in the SOD1G93A transgenic mouse model for ALS. This model does not present any clinical symptoms until postnatal day 90 (P90) (green line). The onset of the symptoms takes place between P90 and P120 (yellow line), followed by progressive paralysis and death until P140 (red line).
Transgenic mice expressing the human wild type SOD1 (hSOD1WT) develop neuronal defects such as mitochondrial swelling and vacuolization, axonal degeneration predominantly in the spinocerebellar tracts, but loss of spinal motor neurons occurs after two years of age [70]. It is worth mentioning that overexpression of hSOD1WT in normal mice at a level similar to that of the mutant SOD1 in transgenic SOD1G93A models has been found sufficient to cause ALS-like syndrome and death at around postnatal day 370 [71]. When hSOD1WT was co-overexpressed with a mutant form of SOD1, such as G93A [70], G85R [72] and A4V [73], it promoted and accelerated the generation of the disease compared to the mutant SOD1 alone.
Toxic gain of function of SOD1 is definitely related to ALS, but the implication of loss of function of SOD1 has been also investigated. Complete deletion of Sod1 from mice did not cause any serious ALS-related effect. In fact, homozygous Sod1 knockout mice develop normally without any motor neuron disruption, and only in late adulthood become vulnerable to motor neuron loss after axonal injury [74]. Furthermore, these mice present additional defects like age-related muscle atrophy [75], distal motor axonopathy [76] and reduced fertility of the females [77] and usually die due to liver cancer [78]. These findings suggest that disease occurrence cannot be attributed to the loss of function of SOD1.
Evaluation of SOD1 Models: Translation in Humans
The SOD1 mouse model is considered as one of the preclinical models for ALS that successfully replicates several patients’ symptoms. Biosamples from patients with ALS, such as cerebrospinal fluid [79] and serum [80] show increased free radical damage. Oxidative damage to RNA species, protein, lipid and DNA has been also detected in SOD1G93A mouse models [81- 83]. miR‑155 is upregulated in the spinal cord and peripheral blood cells of ALS patients leading to an increase of pro-inflammatory cytokine secretion and macrophage inflammatory responses. This result was also found in SOD1G93A mice [84]. In motor cortex and spinal cord of ALS patients, progressive dysfunction and degeneration of gray matter oligodendrocytes has been identified. This feature was replicated in SOD1G93A mouse model as well, as extensive degeneration of gray matter oligodendrocytes in the spinal cord was found prior to disease onset [85]. The loss of glial glutamate transporter (EAAT2/GLT- 1) has been observed in samples from patients with familial or sporadic ALS [86]. This loss of GLT-1 has been reported in SOD1 rodent models of ALS, too [53]. Microglia, the innate immune cells of the nervous system, become activated in ALS [87]. This has been also demonstrated in SOD1G93A mice [88]. It has been found that astrocytes derived from human ALS patients and SOD1G93A mice promote the reduction in expression of major histocompatibility complex class I (MHCI) molecules in motor neurons and render motor neurons vulnerable to astrocyte-induced cell death [89]. Using astrocytes generated from post-mortem tissue of both familial and sporadic ALS patients, Haidet- Phillips and colleagues showed that these are toxic to mouse embryonic stem cell-derived motor neurons [90]. Similarly, rodent astrocytes expressing mutant SOD1 are toxic for mouse primary and embryonic stem cell-derived motor neurons [91].
This demonstrates that ALS is a non-cell autonomous disease and SOD1 mouse models can be used to further examine it.
The two FDA approved drugs for ALS in humans, riluzole and edaravone, have been initially validated in the SOD1G93A mouse model. Riluzole (marketed as Rilutek and Teglutik) was first studied in SOD1G93A mutant mice where it prolonged survival but did not delay disease onset [92]. It was assumed that riluzole is only an antiglutamatergic drug but it has been shown to influence glycine and GABA receptors and neuronal survival as well [93]. Edaravone, (marketed as Radicava and Radicut) a free-radical scavenger used to treat stroke patients in Japan [94] , was tested in SOD1G93A mutant mice and shown to be effective in slowing down symptom progression and motor neurodegeneration [95]. Phase III studies of edaravone were successful [96] and it has been FDA approved since 2017 as a drug for ALS.
Regarding potential therapeutic drugs, mexiletine, an antiarrhythmic, local anaesthetic [97,98], prevented motor neuron death in experiments with SOD1G93A astrocyte conditioned media in wild type primary spinal cultures [99]. Mexiletine was administered to ALS patients and has already reached phase IV [100]. Another compound, vitamin E, which reduces oxidative damage, delayed disease onset in SOD1G93A mice but did not prolong survival [92]. In ALS clinical trials Vitamin E, as an add-on therapy to riluzole, resulted in a marginal trend without reaching significance [101]. Interestingly, as reviewed by Ittner et al., Vitamin E has reached phase III trial [100] which should be completed by now.
Although SOD1 mouse model based research has contributed in drug design and development, evidence in the literature shows that some of the drugs that are effective in SOD1 mice, do not have similar benefits in ALS patients. For instance, ceftriaxone, a GLT-1 translational activator, delayed loss of neurons and increased mouse survival in SOD1G93A mice [102], but did not have any effect in the survival of the ALS patients [103]. In particular, patients treated with ceftriaxone in phase I and phase II presented slower decline but phase III failed to show beneficial effects for the drug. Olesoxime, a molecule with potential neuroprotective properties, showed reduced denervation from 60% to 30%, delayed disease onset and improved survival in SOD1G93A mice [104]. However, as an add-on therapy to riluzole in patients, survival was not improved in phase II–III trials [105]. Although insulin-like growth factor (IgF) prolonged life and delayed disease progression in SOD1G93A mice [106], it does not change patients’ survival [107]. Gabapentin, a GABA analogue, prolonged survival in SOD1G93A mice [92] but didn’t change the muscle strength and survival in human trials [108]. Nowadays, as drug repurposing is conducted, existing approved drugs for different diseases are tested in SOD1 mice in order to detect any beneficial effects for ALS patients. Better results could emerge if potential compounds would be tested in two different transgenic mouse models, such as SOD1 and C9ORF72.
These differences between the effectiveness of the drugs in SOD1 mouse models and in ALS patients could be attributed to many factors. In mice the drug may be administered before the symptoms onset; something that is not feasible in humans because the ALS patients participate in trials only after the diagnosis of the disorder. Another impediment is related to the fact that ALS is a heterogeneous disorder. Patients in clinical trials carry different ALS-related mutations and present different disease onset. This heterogeneity could lead to false negative results for compounds efficient in mice pointing to the need of evaluating the drugs in separate groups of patients. Another factor that may explain the difference in effectiveness of drugs in SOD1 mouse models versus ALS patients is the difference in pharmacodynamics and pharmacokinetics between mice and humans. So, it is uncertain whether the patient would benefit from the successful treatment in mice. Also, confounding variables such as gender, transgene copy number, litter, and exclusion criteria may not be considered in preclinical experiments in SOD1 mice which may lead to false positive results in mice [109]. In addition, uncontrolled variables may lead to false negative results in mice, which means that potential beneficial effects of the drugs are not detected in mice and these compounds never reach ALS patients. Another obstacle in SOD1 mouse models preclinical studies is the fact that this mouse reproduce many symptoms but not all hallmarks of human ALS [110] such as changes in RNA metabolism (which is modelled by TDP43 mice) . Also, anatomical and physiological differences between mice and humans may be responsible for some of the discrepancies in drug effects and should be taken into consideration during study design.
In light of these observations, many solutions have been proposed to solve the differences between preclinical (mouse models) and phase I - III clinical trials (ALS patients). Since ALS is a heterogeneous and multifactorial disease, compound combinations, already found to be effective in mice, may serve as candidate therapeutic strategy for human trials [111]. Attempts following this strategy have already started. For instance, riluzole and sodium phenylbutyrate (NaPB), a histone deacetylase inhibitor which enhances astrocytic neurotrophin synthesis, synergistically increased histone acetylation and extended survival and improved both clinical and neuropathological phenotypes in SOD1G93A mice [112]. In ALS patients, administration of NaPB and riluzole or NaPB alone increased blood histone acetylation levels [113].
Recently, the ALS Therapy Development Institute (TDI) proposed guidelines to reduce the number of false positives in preclinical studies such as reporting all observed changes in animal traits during the course of treatment and variability in death occurrence [110]. In addition, five key points have been suggested to facilitate the transition of treatments from mice to men: the right target, the right patient, the right tissue, the right safety and the right commercial potential [114]. Finally, it has been proposed that the success rate of the clinical trials might be increased if patients are separated in different groups based on mutations and disease onset. Effective compounds in preclinical phase on the SOD1 mouse models, should be tested in all ALS patients but also in a separate cohort carrying only SOD1 mutations. [115].
Conclusion
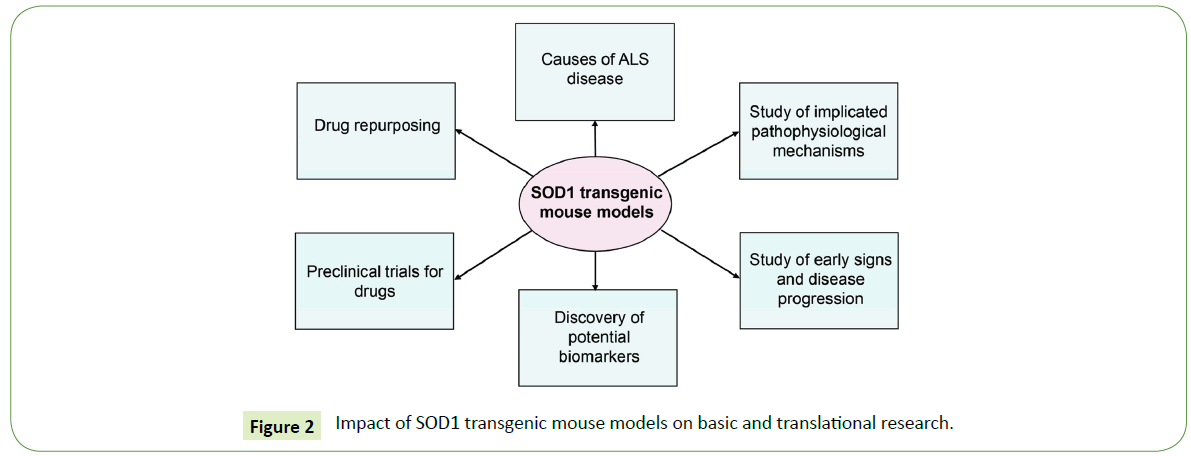
Research on ALS is of high importance for the diagnosis and treatment of this devastating disease. Mechanisms of disease, symptoms onset and disease progression are studied in mouse models for ALS. Collective evidence demonstrates that SOD1 transgenic mice, which were the first generated, recapitulate a large spectrum of the ALS symptoms. SOD1 mouse models have been extensively used in basic and translational research as summarized in Figure 2. In particular, they are studied to unravel causes of the disease and explore many aspects of the mechanisms involved in disease progression. Early signs of the disorder and potential biomarkers identified in these models will enable earlier diagnosis. SOD1 transgenic mice have been also used as preclinical models for the design of drugs. Importantly, although many effective drugs in SOD1 mice are not beneficial or are even toxic for patients, the only two FDA approved drugs for the ALS treatment, riluzole and edaravone, were initially validated in SOD1 mice. Ongoing research on mouse models for ALS will provide the opportunity to design novel genetic and pharmacological approaches for potential therapeutic interventions in humans.
Acknowledgement
We would like to thank Ismini Rozani and Prof. Gareth B. Miles for their constructive and fruitful comments on the manuscript.
Funding
MM and EK are funded by State Scholarships Foundation (IKY), co-financed by the European Union (European Social Fund - ESF) and Greek national funds through the actions entitle “Reinforcement of Doctoral Researchers” and “Reinforcement of Postdoctoral Researchers”, in the framework of the Operational Programme “Human Resources Development Program, Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) 2014 – 2020. LZ is funded by Fondation Santé.
References
- Daube JR (2000) Electrodiagnostic studies in amyotrophic lateral sclerosis and other motor neuron disorders. Muscle Nerve 23: 1488-1502.
- Ferguson TA, Lauren B, Elman LB (2007) Clinical presentation and diagnosis of Amyotrophic Lateral Sclerosis. NeuroRehabilitation 22: 409-416.
- Talbott EO, Malek AM, Lacomis D (2016) The epidemiology of amyotrophic lateral sclerosis. Handb Clin Neurol 138: 225-238.
- Pasinelli P, Brown RH (2006) Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci 7: 710-723.
- Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, et al. (2011) Amyotrophic lateral sclerosis. Lancet 377: 942-955.
- Swinnen B, Robberecht W (2014) The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurosci 10: 661-670.
- Sta M, Sylva-Steenland RM, Casula M, de Jong JM, Troost D, et al. (2011) Innate and adaptive immunity in amyotrophic lateral sclerosis: evidence of complement activation. Neurobiol Dis 42: 211-220.
- Staats KA, Van Den Bosch L (2009) Astrocytes in amyotrophic lateral sclerosis: direct effects on motor neuron survival. J Biol Phys 35: 337-346.
- Al-Chalabi A, Hardiman O (2013) The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol 9: 617-628.
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, et al. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362: 59-62.
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130-133.
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, et al. (2008) TDP- 43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319: 1668-1672.
- Ratti A, Buratti E (2016) Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J Neurochem 138: 95-111.
- Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, et al. (2006) ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet 38: 411-413.
- Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205-1208.
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208-1211.
- Lai SL, Abramzon Y, Schymick JC, Stephan DA, Dunckley T, et al. (2011) FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol Aging 32: 550.e1-4.
- Maruyama H, Morino H, Ito H, Izumi Y, Kato H, et al. (2010) Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465: 223-226.
- Dejesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, et al. (2011) Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9plinked FTD and ALS. Neuron 72: 245-256.
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, et al. (2011) A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72: 257-268.
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, et al. (2011) Mutations in UBQLN2 cause dominant X-linked juvenile and adultonset ALS and ALS/dementia. Nature 477: 211-215.
- Fecto F, Yan J, Vemula SP, Liu E, Yang Y, et al. (2011) SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol 68: 1440-1446.
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, et al. (1994) Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264: 1772-1775.
- Stallings NR, Puttaparthi K, Luther CM, Burns DK, Elliott JL (2010) Progressive motor weakness in transgenic mice expressing human TDP-43. Neurobiol Dis 40: 404-414.
- Xu YF, Gendron TF, Zhang YJ, Lin WL, D'Alton S, et al. (2010) Wild-type human TDP-43 expression causes TDP43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci 30: 10851-10859.
- Arnold ES, Ling SC, Huelga SC, Lagier-Tourenne C, Polymenidou M, et al. (2013) ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci USA 110: E736-745.
- Mitchell JC, Constable R, So E, Vance C, Scotter E, et al. (2015) Wild type human TDP-43 potentiates ALS-linked mutant TDP-43 driven progressive motor and cortical neuron degeneration with pathological features of ALS. Acta Neuropathol Commun 3: 36.
- Xu YF, Zhang YJ, Lin WL, Cao X, Stetler C, et al. (2011) Expression of mutant TDP-43 induces neuronal dysfunction in transgenic mice. Mol Neurodegener 6: 73.
- Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH (2009) TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA 106: 18809-18814.
- Borchelt DR, Sisodia SS (1996) Loss of functional prion protein: a role in prion disorders? Chem Biol 3: 619-621.
- Wils H, Kleinberger G, Janssens J, Pereson S, Joris G, et al. (2010) TDP- 43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA 107: 3858-3863.
- Shan X, Chiang PM, Price DL, Wong PC (2010) Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci USA 107: 16325-16330.
- Vidal M, Morris R, Grosveld F, Spanopoulou E (1990) Tissue-specific control elements of the Thy-1 gene. Embo J 9: 833-840.
- Caroni P (1997) Overexpression of growth-associated proteins in the neurons of adult transgenic mice. J Neurosci Methods 71: 3-9.
- Tsai KJ, Yang CH, Fang YH, Cho KH, Chien WL, et al. (2010) Elevated expression of TDP-43 in the forebrain of mice is sufficient to cause neurological and pathological phenotypes mimicking FTLD-U. J Exp Med 207: 1661-1673.
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, et al. (1996) Control of memory formation through regulated expression of a CaMKII transgene. Science 274: 1678-1683.
- Kraemer BC, Schuck T, Wheeler JM, Robinson LC, Trojanowski JQ, et al. (2010) Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol 119: 409-419.
- Sephton CF, Good SK, Atkin S, Dewey CM, Mayer PIII, et al. (2010) TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem 285: 6826-6834.
- Wu LS, Cheng WC, Hou SC, Yan YT, Jiang ST, et al. (2010) TDP- 43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis 48: 56-62.
- Chiang PM, Ling J, Jeong YH, Price DL, Aja SM, et al. (2010) Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci U S A 14: 16320-16324.
- Hicks GG, Singh N, Nashabi A, Mai S, Bozek G, et al. (2000) Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat Genet 24: 175-179.
- Kino Y, Washizu C, Kurosawa M, Yamada M, Miyazaki H, et al. (2015) FUS/TLS deficiency causes behavioral and pathological abnormalities distinct from amyotrophic lateral sclerosis. Acta Neuropathol Commun 3: 24.
- Mitchell JC, McGoldrick P, Vance C, Hortobagyi T, Sreedharan J, et al. (2013) Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age and dose-dependent fashion. Acta Neuropathol 125: 273-88.
- Sharma A, Lyashchenko AK, Lu L, Nasrabady SE, Elmaleh M, et al. (2016) ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat Commun 7: 10465.
- Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, et al. (2015) Neurodegeneration C9ORF72 repeat expansions in mice cause TDP- 43 pathology, neuronal loss, and behavioral deficits. Science 348: 1151-1154.
- Peters OM, Cabrera GT, Tran H, Gendron TF, McKeon JE, et al. (2015) Human C9ORF72 hexanucleotide expansion reproduces RNA foci and dipeptide repeat proteins but not neurodegeneration in BAC transgenic mice. Neuron 88: 902-909.
- O'Rourke JG, Bogdanik L, Muhammad AK, Gendron TF, Kim KJ, et al. (2005) C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron 88: 892-901.
- Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, et al. (2016) Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC containing RNAs. Neuron 90: 535-550.
- Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, et al. (2016) C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90: 521-534.
- Koppers M, Blokhuis AM, Westeneng HJ, Terpstra ML, Zundel CA, et al. (2015) C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann Neurol 78: 426-438.
- Sudria-Lopez E, Koppers M, de Wit M, van der Meer C, Westeneng HJ, et al. (2016) Full ablation of C9orf72 in mice causes immune system-related pathology and neoplastic events but no motor neuron defects. Acta Neuropathol 132: 145-147.
- Gorrie GH, Fecto F, Radzicki D, Weiss C, Shi Y, et al. (2014) Dendritic spinopathy in transgenic mice expressing ALS/dementia-linked mutant UBQLN2. Proc Natl Acad Sci USA 111: 14524-14529.
- Bruijn LI, Becher MW, Lee MK, Anderson KL, Jenkins NA, et al. (1997) ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron 18: 327-338.
- Pramatarova A, Laganiere J, Roussel J, Brisebois K, Rouleau GA (2001) Neuron-specific expression of mutant superoxide dismutase 1 in transgenic mice does not lead to motor impairment. J Neurosci 21: 3369-3374.
- Lino MM, Schneider C, Caroni P (2002) Accumulation of SOD1 mutants in postnatal motoneurons does not cause motoneuron pathology or motoneuron disease. J Neurosci 22: 4825-4832.
- Gong YH, Parsadanian AS, Andreeva A, Snider WD, Elliott JL (2000) Restricted expression of G86R Cu/Zn superoxide dismutase in astrocytes results in astrocytosis but does not cause motoneuron degeneration. J Neurosci 20: 660-665.
- Turner BJ, Ackerley S, Davies KE, Talbot K (2010) Dismutasecompetent SOD1 mutant accumulation in myelinating Schwann cells is not detrimental to normal or transgenic ALS model mice. Hum Mol Genet 19: 815-824.
- Pfohl SR, Halicek MT, Mitchell CS (2015) Characterization of the contribution of genetic background and gender to disease progression in the SOD1 G93A mouse model of amyotrophic lateral sclerosis: a meta-analysis. J Neuromuscul Dis 2: 137-150.
- Dal Canto MC, Gurney ME (1995) Neuropathological changes in two lines of mice carrying a transgene for mutant human Cu, Zn SOD, and in mice overexpressing wild type human SOD: a model of familial amyotrophic lateral sclerosis (FALS). Brain Res 676: 25-40.
- Dal Canto MC, Gurney ME (1997) A low expressor line of transgenic mice carrying a mutant human Cu, Zn superoxide dismutase (SOD1) gene develops pathological changes that most closely resemble those in human amyotrophic lateral sclerosis. Acta Neuropathol 93: 537-550.
- Xu Z, Jung C, Higgins C, Levine J, Kong J (2004) Mitochondrial degeneration in amyotrophic lateral sclerosis. J Bioenerg Biomembr 36: 395-399.
- Higgins CMJ, Jung C, Xu Z (2003) ALS-associated mutant SOD1G93A causes mitochondrial vacuolation by expansion of the intermembrane space and by involvement of SOD1 aggregation and peroxisomes. BMC Neurosci 4: 16.
- Casoni F, Basso M, Massignan T, Gianazza E, Cheroni C, et al. (2005) Protein nitration in a mouse model of familial amyotrophic lateral sclerosis: possible multifunctional role in the pathogenesis. J Biol Chem 280: 16295-16304.
- Pardo AC, Wong V, Benson LM, Dykes M, Tanaka K, et al. (2006) Loss of the astrocyte glutamate transporter GTL1 modifies disease in SOD(G39A) mice. Exp Neurol 201: 120-130.
- Sasaki S, Warita H, Abe K, Iwata M (2005) Impairment of axonal transport in the axon hillock and the initial segment of anterior horn neurons in transgenic mice with a G93A mutant SOD1 gene. Acta Neuropathol 110: 48-56.
- Kennel PF, Finiels F, Revah F, Mallet J (1996) Neuromuscular function impairment is not caused by motor neurone loss in FALS mice: an electromyographic study. Neuroreport 7: 1427-1431.
- Frey D, Schneider C, Xu L, Borg J, Spooren W, et al. (2000) Early and selective loss of neuromuscular synapse subtypes with low sprouting competence in motoneuron diseases. J Neurosci 20: 2534-2542.
- Basso M, Massignan T, Samengo G, Cheroni C, De Biasi S, et al. (2006) Insoluble mutant SOD1 is partly oligoubiquitinated in amyotrophic lateral sclerosis mice. J Biol Chem 281: 33325-33335.
- Philips T, Rothstein JD (2014) Glial cells in amyotrophic lateral sclerosis. Exp Neurol 262 Pt B: 111-120.
- Jaarsma D, Haasdijk ED, Grashorn JA, Hawkins R, van Duijn W, et al. (2000) Human Cu/Zn superoxide dismutase (SOD1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant SOD1. Neurobiol Dis 7:623-643.
- Graffmo KS, Forsberg K, Bergh J, Birve A, Zetterstrom P, et al. (2013) Expression of wild-type human superoxide dismutase-1 in mice causes amyotrophic lateral sclerosis. Hum Mol Genet 22:51-60.
- Wang L, Deng HX, Grisotti G, Zhai H, Siddique T, et al. (2009) Wildtype SOD1 overexpression accelerates disease onset of a G85R SOD1 mouse. Hum Mol Genet 18:1642-1651.
- Deng HX, Shi Y, Furukawa Y, Zhai H, Fu R, et al. (2006) Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc Natl Acad Sci USA 103:7142-7147.
- Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, et al. (1996) Motor neurons in Cu/Zn superoxide dismutasedeficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet 13: 43-47.
- Muller FL, Song W, Liu Y, Chaudhuri A, Pieke-Dahl S, et al. (2006) Absence of CuZn superoxide dismutase leads to elevated oxidative stress and acceleration of age-dependent skeletal muscle atrophy. Free Radic Biol Med 274: 1678-1683.
- Shefner JM, Reaume AG, Flood DG, Scott RW, Kowall NW, et al. (1999) Mice lacking cytosolic copper/zinc superoxide dismutase display a distinctive motor axonopathy. Neurology 53: 1239-1246.
- Ho YS, Gargano M, Cao J, Bronson RT, Heimler I, et al. (1998) Reduced fertility in female mice lacking copper-zinc superoxide dismutase. J Biol Chem 273: 7765-7769.
- Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, et al. (2005) CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene 24: 367-380.
- Smith RG, Henry YK, Mattson MP, Appel SH (1998) Presence of 4-hydroxynonenal in cerebrospinal fluid of patients with sporadic amyotrophic lateral sclerosis. Ann Neurol 44: 696-699.
- Simpson EP, Henry YK, Henkel JS, Smith RG, Appel SH (2004) Increased lipid peroxidation in sera of ALS patients: a potential biomarker of disease burden. Neurology 62: 1758-1765.
- Poon HF, Hensley K, Thongboonkerd V, Merchant ML, Lynn BC, et al. (2005) Redox proteomics analysis of oxidatively modified proteins in G93A-SOD1 transgenic mice-a model of familial amyotrophic lateral sclerosis. Free Radic Biol Med 39: 453-462.
- Barber SC, Mead RJ, Shaw PJ (2006) Oxidative stress in ALS: a mechanism of neurodegeneration and a therapeutic target. Biochim Biophys Acta 1762: 1051-1067.
- Chang Y, Kong Q, Shan X, Tian G, Ilieva H, et al. (2008) Messenger RNA oxidation occurs early in disease pathogenesis and promotes motor neuron degeneration in ALS. PLoS One 3: e2849.
- Koval ED, Shaner C, Zhang P, du Maine X, Fischer K, et al. (2013). Method for widespread microRNA‑155 inhibition prolongs survival in ALS-model mice. Hum Mol Genet 22: 4127-4135.
- Kang SH, Fukaya M, Lorenzini I, Cleveland DW, Ostrow LW, et al. (2013) Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat Neurosci 16: 571-579.
- Rothstein JD, Van Kammen M, Levey AI, Martin LJ, Kuncl RW (1995) Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. Ann Neurol 38: 73-84.
- Taylor JP, Brownm RHJr, Cleveland DW (2016) Decoding ALS: from genes to mechanism. Nature 539: 197-206.
- Hall ED, Oostveen JA, Gurney ME (1998) Relationship of microglial and astrocytic activation to disease onset and progression in a transgenic model of familial ALS. Glia 23: 249-256.
- Song S, Miranda CJ, Braun L, Meyer K, Frakes A, et al. (2016) Major histocompatibility complex class I molecules protect motor neurons from astrocyte-induced toxicity in amyotrophic lateral sclerosis. Nat Med 22: 397-403.
- Haidet-Phillips AM, Hester ME, Miranda CJ, Meyer K, Braun L, et al. (2011) Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol 29: 824-828.
- Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, et al. (2007) Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci 10: 615-622.
- Gurney ME, Cutting FB, Zhai P, Doble A, Taylor CP, et al. (1996) Benefit of vitamin E, riluzole, and gabapentin in a transgenic model of familial amyotrophic lateral sclerosis. Ann Neurol 39: 147-157.
- Bellingham MC (2011) A review of the neural mechanisms of action and clinical efficiency of riluzole in treating amyotrophic lateral sclerosis: what have we learned in the last decade? CNS Neurosci Ther 17: 4-31.
- Edaravone Acute Infarction Study Group (2003) Effect of a novel free radical scavenger, edaravone (MCI-186), on acute brain infarction: randomized, placebo-controlled, double-blind study at multicenters. Cerebrovasc Dis 15: 222-229.
- Ito H, Wate R, Zhang J, Ohnishi S, Kaneko S, et al. (2008) Treatment with edaravone, initiated at symptom onset, slows motor decline and decreases SOD1 deposition in ALS mice. Exp Neurol 213: 448-455.
- Writing Group; Edaravone (MCI-186) ALS 19 Study Group (2017) Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebocontrolled trial. Lancet Neurol 16: 505-512.
- Schrader BJ, Bauman JL (1986) Mexiletine: a new type I antiarrhythmic agent. Drug Intell Clin Pharm 20: 255-260.
- Olschewski A, Schnoebel-Ehehalt R, Li Y, Tang B, Bräu ME, et al. (2009) Mexiletine and lidocaine suppress the excitability of dorsal horn neurons. Anesth Analg 109: 258-264.
- Fritz E, Izaurieta P, Weiss A, Mir FR, Rojas P, et al. (2013) Mutant SOD1-expressing astrocytes release toxic factors that trigger motor neuron death by inducing hyper-excitability. J Neurophysiol 109: 2803-2814.
- Ittner LM, Halliday GM, Kril JJ, Gotz J, Hodges JR, et al. (2015) FTD and ALS-translating mouse studies into clinical trials. Nat Rev Neurol 11: 360-366.
- Graf M, Ecker D, Horowski R, Kramer B, Riederer P, et al. (2005) High dose vitamin E therapy in amyotrophic lateral sclerosis as addon therapy to riluzole: results of a placebo-controlled double-blind study. J Neural Transm (Vienna) 112: 649-660.
- Rothstein JD, Patel S, Regan MR, Haenggeli C, Huang YH, et al. (2005) Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature 433: 73-77.
- Cudkowicz ME, Titus S, Kearney M, Yu H, Sherman A, et al. (2014) Safety and efficacy of ceftriaxone for amyotrophic lateral sclerosis: a multi-stage, randomised, double-blind, placebo-controlled trial. Lancet Neurol 13: 1083-1091.
- Sunyach C, Michaud M, Arnoux T, Bernard-Marissal N, Aebischer J, et al. (2012) Olesoxime delays muscle denervation, astrogliosis, microglial activation and motoneuron death in an ALS mouse model. Neuropharmacology 62: 2346-2352.
- Lenglet T, Lacomblez L, Abitbol JL, Ludolph A, Mora JS, et al. (2014) A phase II–III trial of olesoxime in subjects with amyotrophic lateral sclerosis. Eur J Neurol 21: 529-536.
- Kaspar BK, Lladó J, Sherkat N, Rothstein JD, Gage FH (2003) Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science 301: 839-842.
- Sorenson EJ, Windbank AJ, Mandrekar JN, Bamlet WR, Appel SH, et al. (2008) Subcutaneous IGF‑1 is not beneficial in 2‑year ALS trial. Neurology 71: 1770-1775.
- Miller RG, Moore DH, Gelinas DF, Dronsky V, Mendoza M, et al. (2001) Phase III randomized trial of gabapentin in patients with amyotrophic lateral sclerosis. Neurology 56: 843-848.
- Scott S, Kranz JE, Cole J, Lincecum JM, Thompson K, et al.(2008) Design, power, and interpretation of studies in the standard murine model of ALS. Amyotroph Lateral Scler 9: 4-15.
- Perrin S (2014) Preclinical research: make mouse studies work. Nature 507: 423-425.
- Kong Q, Carothers S, Chang Y, Glenn Lin CL (2012) The importance of preclinical trial timing - a potential reason for the disconnect between mouse studies and human clinical trials in ALS. CNS Neurosci Ther 18: 791-793.
- Del Signore SJ, Amante DJ, Kim J, Stack EC, Goodrich S, et al. (2009) Combined riluzole and sodium phenylbutyrate therapy in transgenic amyotrophic lateral sclerosis mice. Amyotroph Lateral Scler 10: 85-94.
- Cudkowicz ME, Andres PL, Macdonald SA, Bedlack RS, Choudry R, et al. (2009) Phase 2 study of sodium phenylbutyrate in ALS. Amyotroph Lateral Scler 10: 99-106.
- Cook D, Brown D, Alexander R, March R, Morgan P, et al. (2014) Lessons learned from the fate of AstraZeneca’s drug pipeline: a fivedimensional framework. Nat Rev Drug Discov 13: 419-431.
- Benatar M, Wuu J, Andersen PM, Atassi N, David W, et al. (2018) Randomized, double blind, placebo-controlled trial of arimoclom. Neurology 90: e565-e574.
- Wong PC, Pardo CA, Borchelt DR, Lee MK, Copeland NG, et al. (1995) An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron 14: 1105-1116.
- Ripps ME, Huntley GW, Hof PR, Morrison JH, Gordon JW (1995) Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 92: 689-693.
- Tobisawa S, Hozumi Y, Arawaka S, Koyama S, Wada M, et al. (2003) Mutant SOD1 linked to familial amyotrophic lateral sclerosis, but not wild-type SOD1, induces ER stress in COS7 cells and transgenic mice. Biochem Biophys Res Commun 303: 496-503.
- Jonsson PA, Ernhill K, Andersen PM, Bergemalm D, Brannstrom T, et al. (2004) Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain 127: 73-88.
- Chang-Hong R, Wada M, Koyama S, Kimura H, Arawaka S, et al. (2005) Neuroprotective effect of oxidized galectin-1 in a transgenic mouse model of amyotrophic lateral sclerosis. Exp Neurol 194: 203-211.
- Jonsson PA, Graffmo KS, Brannstrom T, Nilsson P, Andersen PM, et al. (2006) Motor neuron disease in mice expressing the wild type-like D90A mutant superoxide dismutase-1. J Neuropathol Exp Neurol 65: 1126-1136.
- Wang J, Xu G, Li H, Gonzales V, Fromholt D, et al. (2005) Somatodendritic accumulation of misfolded SOD1-L126Z in motor neurons mediates degeneration: alphaB-crystallin modulates aggregation. Hum Mol Genet 14: 2335-2347.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences