VDAC Differential Interactome in Chicken Brain: Possible Hints to an Intrinsically Distinct Metabolism
Marcelo de Cerqueira Cesar
Carla Rossini Crepaldi1, Helen Julie Laure2, Jose Cesar Rosa2 and Marcelo de Cerqueira Cesar1*
1Department of Basic Sciences, School of Animal Science and Food Engineering, University of Sao Paulo, Pirassununga, Brazil
2Protein Chemistry Center and Department of Molecular and Cellular Biology and Pathogenic Bio agents, Faculty of Medicine of Ribeirao Preto, University of Sao Paulo, Sao Paulo, Brazil
- *Corresponding Author:
- Marcelo de Cerqueira Cesar
Laboratory of Neuroscience and Proteomics
School of Animal Science and Food Engineering
University of Sao Paulo, Av. Duque de Caxias Norte 225, 13635-900, Pirassununga, SP, Brazil
Tel: 19 35654095
E-mail: mccesar@usp.br
Received date:February 18, 2016; Accepted date: March 23, 2016; Published date: March 30, 2016
Citation: Crepaldi CR, Laure HJ, Rosa JC, et al. VDAC Differential Interactome in Chicken Brain: Possible Hints to an Intrinsically Distinct Metabolism. J Transl Neurosci. 2016, 1:1.
Copyright: © 2016 Crepaldi CR, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
The kinetic assembly of protein complexes containing the VDAC (voltage dependent anion channel) follows a quite different pattern in bovine brain in comparison with rat neuronal cells. In order to investigate if the differential expression of VDACs between chicken brain when compared with bovine and rat brain mitochondria was linked to a differential VDAC interactome, we utilized BN-PAGE of mitochondria treated with dodecyl maltoside, followed by a seconddimensional SDS-PAGE. Unusually perhaps, several VDAC interactant proteins in chicken brain were not members of VDAC interactome in rat and bovine brain, which possibly suggests that these cells exhibit intrinsically differential responses to events such as neuronal cell death, bioenergetics, oxygen consumption and oxidative insults.
Keywords
Hexokinase; VDAC; Interactome; BN / SDS PAGE; Mitochondria; Chicken; Brain
Introduction
Hexokinase (HK, EC 2.7.1.1) mediates the first step of glucose catabolism, phosphorylating glucose to produce glucose 6-phosphate. The regulation of HK activity plays a major role in governing the rate of cerebral Glc utilization while avoiding production of neurotoxic lactate [1,2]. The Type I isozyme of hexokinase (HK 1) accounts for more than 90% of the total hexokinase activity in mammalian brain, where it exists predominantly in a mitochondrially bound form [2].
HK I attaches to the voltage dependent anion channel (VDAC) via their N-terminal region facilitating access to intra-mitochondrial ATP. It has been suggested that binding of HK modulates VDAC’s role in apoptosis [3]. VDAC proteins constitute the major pathway for metabolic exchange across the mitochondrial outer membrane (MOM). Two isoforms of VDAC, i.e., VDAC1 and VDAC2 are known to be expressed ubiquitously in chicken brain mitochondria [4].
VDAC1 regulates metabolic and energetic functions of mitochondria; its down-expression should affect cell metabolism and normal mitochondrial function [5]. The VDAC isoform 2 has anti-apoptotic roles as a specific inhibitor of BAK-dependent mitochondrial apoptosis [6]. Changes in the levels of VDAC1 and VDAC2 expression were observed under various pathological conditions [7]. It is interesting to note that cells with low levels of VDAC1 showed 4-fold-lower ATP-synthesis capacity and contained low ATP and ADP levels. In these cells there was a strong correlation between ATP levels and cell growth, suggesting limited metabolite exchange between mitochondria and cytosol [8].
It has been studied the possibility that differences in the relative expression of VDAC isoforms could be a factor in determining the differences in species-dependent ratio of hexokinase binding sites on bovine, avian and rat brain mitochondria. In this research, VDAC1 was the most abundantly expressed of the three isoforms. Moreover, chicken brain mitochondria showed the highest VDAC1 expression and the lowest VDAC2 levels between the three species [4]. The same phenomenon, increase of VDAC1 and decrease of VDAC2 has been detected in pharmaco-resistant epilepsy [9].
There are a variety of conventional (liquid chromatography, ultracentrifugation, and sucrose density gradient centrifugation) and nonconventional methods (co-immunoprecipitation, epitope-tagging, tandem affinity purification, and GST-pull-down) available for isolation of multiprotein complexes. However, most of these techniques often separate a population of such assemblies. To isolate individual complexes, further separation is required, which can be achieved by two-dimensional blue native SDS-PAGE [10]. In BN-PAGE, the Coomassie Brilliant Blue G-250 dye is added into the electrophoresis buffer. This anionic dye binds to the surface of the membrane proteins and facilitates their migration in the native polyacrylamide gel [11]. As a result, distinct protein complexes are separated due the sieving effect of the polyacrylamide gel, but protein – protein interactions among the multiprotein complex subunits are still retained. Although initially developed for the separation of mitochondrial and chloroplast membrane proteins, recent contributions of this approach have been made in studies of protein complexes in termophilic (Clostridium termocellum), antibiotic productive (Streptomyces coelicolor), sulfate-reducing bacteria and in pathogens associated with chronic periodontitis in humans [11-15]. BN / SDS-PAGE have also been used to study protein complexes in a variety of tissues, such as erythrocytes, rat muscle, colorectal cancer and bladder epithelial cells [10,12,16,17].
Recent studies indicate that the kinetic assembly of protein complexes containing the VDAC follows a pattern quite different in bovine and rat brain [18]. In order to investigate if the differential expression of VDACs between chicken brain when compared with bovine and rat brain mitochondria was linked to a differential VDAC interactome, we utilized BN-PAGE of mitochondria treated with dodecyl maltoside. After BN-PAGE, a second-dimensional SDS-PAGE was performed to separate polypeptides as components of VDAC complexes.
Materials and Method
Preparation of brain mitochondria
Chicken brains were obtained from the school slaughterhouse located on campus. Mitochondria were isolated from avian brain as described previously [2]. Mitochondrial protein concentrations were determined by the Bicinchoninic Acid Method, using the assay kit from Thermo Scientific (Rockford, IL, USA) with bovine serum albumin as standard.
Sample preparation
To determine the optimal conditions for the solubilization of VDAC protein complexes, a series of five different concentrations of dodecyl-maltoside (DDM) (0.25, 0.5, 0.75, 1.0 and 1.5%) were evaluated. The solubilization with 1.0% (w/v) was found to be the most effective. Mitochondrial membranes were solubilized as previously reported [18]. The detergent extracts of mitochondria containing approximately 200 μg of protein before solubilization, were loaded per lane.
2D – BN / Tricine SDS PAGE
BN PAGE was performed with linear 6% - 13% gradient gels, overlaid with a 4% stacking gel [19]. GE Healthcare HMW-Native protein markers (GE Healthcare, Buckinghamshire, UK) were used. For a second dimension, BN-PAGE gel lanes containing the proteins of interest were excised and incubated for 30 min. in equilibrating buffer A, containing 12,5 mM Tris (pH 6.8), 4% SDS, 20% Glycerol and 9% β-mercaptoethanol. The lanes were then dipped into equilibrating buffer B supplemented with 12,5 mM Tris HCl pH 6.8, 4% SDS, 20% Glycerol, and 2.5% Iodoacetamide for 15 min. at room temperature.
A 10% Tricine SDS-PAGE was utilized for the separation of proteins [20]. Proteins were subjected to Coomassie Blue R-250 staining or immunoblotting, to detect the complexes in which VDACs 1 and 2 were found.
Each stained gel was digitized and processed using the ImageQuantTL Capture software system (GE Healthcare). This software contains a graph that shows the intensity at each point along the length of the current lane. This is a threshold parameter, which discards peaks under a certain size (30) in relation to the highest peak on the gel. The higher the percentage value entered here the fewer the peaks likely to be detected in the profile. The sizes of the peaks were calculated after background subtraction.
Protein detection by western blotting
Proteins separated as described above were transferred to nitrocellulose membranes using TE62 Transfer Unit (GE Halthcare) and the buffer system of [21]. Membranes were blocked by incubation (overnight, 4°C) with 5% non-fat dried milk in TBS (20 mM Tris - 0.5 M NaCl, pH 7.5). Blots were probed with suitable dilutions of primary antibodies, and then washed extensively with TBS containing 0.1% (v/v) Tween 20, and incubated with 1:10,000-15,000 dilutions of horseradish peroxidase conjugated secondary antibodies; both primary and secondary antibodies were diluted in TBS containing 1% (w/v) gelatin. After further washing with TBS -Tween, chemi-luminescence was developed using the Super Signal West Pico system from Thermo Scientific (Rockford, IL, USA).
Mass spectrometry and protein identification
The second dimension gel bands were selected and proteins were identified by MALDI-TOF-TOF mass spectrometer after in gel trypsin digestion. Briefly, selected gel bands were excised and combined. SDS and CBB were removed by washing the gels three times with 50% ACN in 0.1 M ammonium bicarbonate, pH 7.8, followed by dehydration in neat ACN. Gel bands were dried in a Speed Vac instrument (Savant, New York, NY) and were swollen in 20 mL of 0.5 mg trypsin (Promega, Madison, USA) in 0.1 M ammonium bicarbonate, pH 7.8, followed by the addition of 50 mL of 0.1 M ammonium bicarbonate to cover the entire gel piece. Trypsin hydrolysis was carried out at 37°C for 24 h and the reaction was stopped by the addition of 5 mL of neat formic acid. Peptides were extracted from gel pieces and desalted in microtips filled with POROS R2 (PerSeptive Biosystems, Foster City, CA) previously equilibrated in 0.2% formic acid. After loading, the sample was desalted with two washes of 150 mL 0.2% formic acid. Peptides were eluted from the microtips with 30 mL of 60% methanol / 5% formic acid. The sample was concentrated, and samples were mixed with matrix solution (5 mg/mL α-cyano-4 hydroxycinnamic acid in 50% acetonitrile / 0.1% trifluoroacetic acid), applied on the MALDI target plate and air dried at room temperature. MALDI-TOF-TOF-MS instrument (Axima Performance, Kratos-Shimadzu, Manchester, UK) was calibrated with a mixture of bradykynin fragment, angiotensin II, renin and ACTH (mass accuracy < 50 ppm). The spectra of CID-MS / MS of each gel band were obtained in data dependent acquisition mode. The peak list was obtained from CID-MS / MS spectra using Launchpad v. 2.8 (Kratos-Shimadzu, Manchester, UK) and submitted to a database search using MASCOT version 2.2.04 against NCBI database version 52.9 selected for taxonomy filter of Gallus gallus. The database search parameters accept one missing trypsin cleavage, carbamidomethylation and methionine oxidation. Mass tolerance was 1.2 Da for precursor ions and 0.8 Da for product ions. Protein was considered to be identified by MASCOT for proteins corresponded to a level of significance of p < 0.05 and FDR less than 0.2%.
Results and Discussion
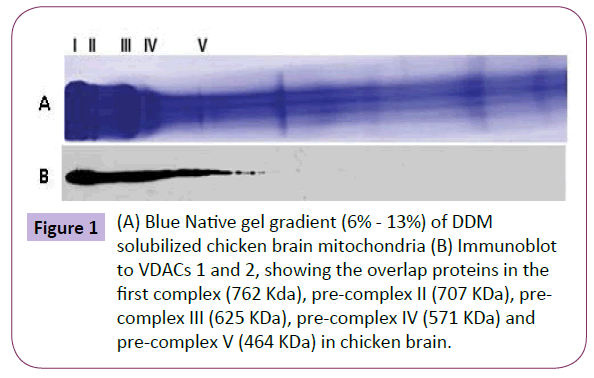
To analyze proteins interacting with VDAC, BN / SDS-PAGE was performed. The VDACs 1 and 2 associated protein complexes were revealed by antibody detection and confirmed by mass spectrometry. In the present paper, we utilized a well-characterized antibody against VDAC1, which was first raised by Poleti et al., which is highly specific for their desired targets [4]. However, the antibody against VDAC2 showed cross-immunoreactivity with VDAC1 from chick brain mitochondria. VDACs were detected in four pre-complexes (II, III, IV and V) and one complex (I) in avian brain mitochondria (Figure 1). The apparent masses of complex I and pre-complexes II, III, IV and V were 762, 707, 625, 571 and 464 kDa, respectively.
Figure 1: (A) Blue Native gel gradient (6% - 13%) of DDM solubilized chicken brain mitochondria (B) Immunoblot to VDACs 1 and 2, showing the overlap proteins in the first complex (762 Kda), pre-complex II (707 KDa), pre-complex III (625 KDa), pre-complex IV (571 KDa) and pre-complex V (464 KDa) in chicken brain.
The general interpretative rule for the analysis of the BN / SDSPAGE is that all protein spots which are situated vertically in the gel are potential subunits of a same protein complex. A second rule is that if it is combined information about the subunit composition of the protein complexes (vertical) and the molecular mass of the complexes (horizontal) and is assumed that the molecular mass of a protein complex is increasing during assembly of the structural subunits of this complex, the analysis of the protein pattern after BN / SDS-PAGE resolves the direction of the stepwise subunit assembly from the lower towards the higher molecular mass of the protein complexes [22].
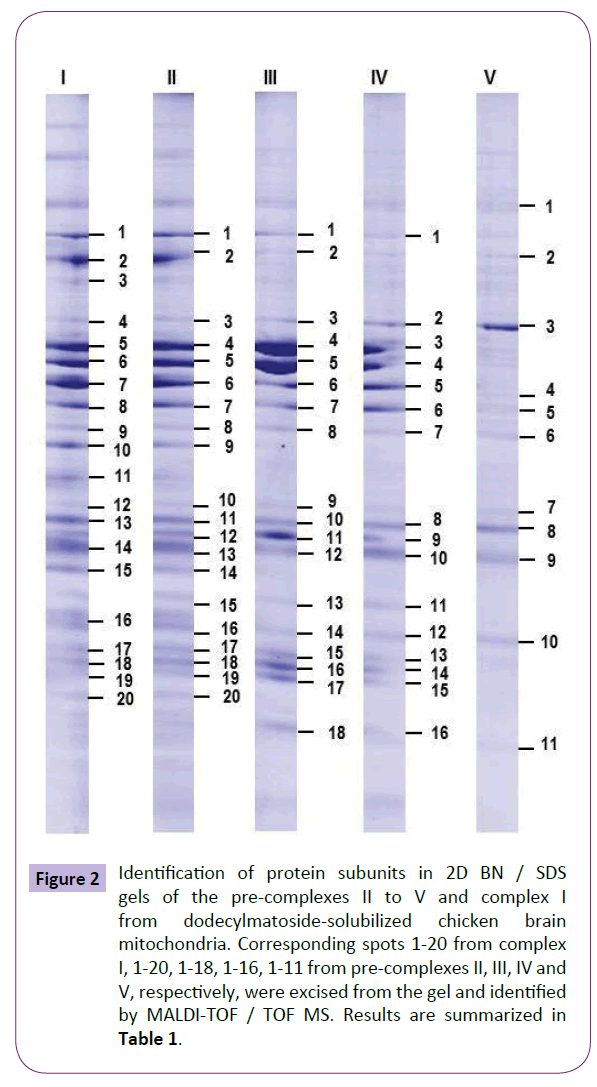
BN / SDS-PAGE were performed on five replicates of five different mitochondrial preparations (each preparation extracted from 3 adult chickens). The analysis of the representative gel from Figure 2 detected 85 spots of VDAC interacting proteins. Among these spots, 54 different proteins were identified with a MASCOT score above the threshold to validate MS data, i.e., 32. The identification of subunits of individual OXPHOs complexes, as VDAC interacting proteins in chicken brain, confirmed the validity of our MS approach. These interactants represent a large variety of functions such as components of TCA cycle, ketone body’s metabolism, scavengers of reactive oxygen species, members of mitochondrial permeability transition pore and cell proliferation between many others.
Figure 2: Identification of protein subunits in 2D BN / SDS gels of the pre-complexes II to V and complex I from dodecylmatoside-solubilized chicken brain mitochondria. Corresponding spots 1-20 from complex I, 1-20, 1-18, 1-16, 1-11 from pre-complexes II, III, IV and V, respectively, were excised from the gel and identified by MALDI-TOF / TOF MS. Results are summarized in Table 1.
The identified proteins and their corresponding MASCOT score, sequence coverage, and number of matched MS / MS are listed in Tables 1-6. The four pre-complexes and the major complex from chicken brain mitochondria can be observed in Figure 2 and Table 1.
| I | II | III | IV | V |
|---|---|---|---|---|
| 3CWB_D | 3CWB_A | 3CWB_A | ANT | |
| ANTI | ANT1 | ANT1 | ANT1 | ANT1 |
| ANT3 | ANT3 | ANT3 | ANT3 | ANT3 |
| AATM | AATM | AATM | AATM | |
| ACON | ACON | |||
| ATPG | ATPG | ATPG | ||
| AT5F1 | AT5F1 | AT5F1 | AT5F1 | |
| ATP5H | ATP5H | ATP5H | ATP5H | |
| ATP5O | ATP5O | ATP5O | ATP5O | |
| ATPB | ATPA | ATPA | ATPA | |
| ATPA | ATPB | ATPB | ATPB | AL1I2 |
| ARALAR2 | ATPD | ATPD | ||
| avANT | avANT | avANT | avANT | avANT |
| BDH1 | BDH1 | BDH1 | ||
| CH60 | CH60 | CH60 | CH60 | CH60 |
| CHPF2 | ||||
| CMBL | CMBL | |||
| COXII | COXII | COXII | COXII | COXII |
| COX4I1 | COX4I1 | COX4I1 | ||
| DOCK2 | DESM | |||
| E1C825 | HADHA | HADHA | ||
| IMMT | IMMT | IMMT | IMMT | |
| QN1 | ||||
| MRP1 | MRP1 | |||
| MDH2 | MDH2 | MDH2 | ||
| NDUA9 | MYH11 | |||
| NDUAA | NDUAA | MPCP | ||
| NDUFA8 | NDUFA8 | NDUFA8 | ||
| NDUFB4 | ||||
| NDUFA12 | NDUFA12 | |||
| NDUFB8 | NDUFB8 | |||
| NDUFB10 | NDUFB10 | NDUFB10 | ||
| NDUFS7 | NDUFS7 | NDUFS7 | ||
| NDUFS8 | NDUFS8 | |||
| NDUFS1 | NDUFS1 | NDUFS1 | ||
| NDUFS3 | NDUFS3 | |||
| NDUV2 | NDUV2 | NDUV2 | ||
| PHB | PHB | PRDX3 | PRDX3 | PRDX3 |
| PHB2 | PHB2 | RBBP8 | RBBP8 | |
| QCRC1 | QCRC1 | |||
| QCRC2 | QCRC2 | QCRC2 | QCRC2 | QCRC2 |
| VDAC2 | VDAC2 | VDAC2 | VDAC2 | VDAC2 |
| XIRP1 | ||||
| WWC2 |
Table 1: Identification results of VDAC-associated protein complex (I) and pre-complexes (II to V) in chicken brain mitochondria.
| Complex I | ||||||||
|---|---|---|---|---|---|---|---|---|
| Spot | Accession NCBI | Name of protein | Function | Mascot score | Sequence coverage (%) | Matches MS/MS | Theoretical | |
| pI | Mr | |||||||
| 1 | gi|57530041 | Mitochondrial inner membrane protein | Other functions | 578 | 13,2 | 14 | 5,72 | 79249 |
| 2 | gi|57529753 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | Oxidative phosphorylation | 1024 | 18,7 | 16 | 6,5 | 79576 |
| 3 | gi|61098440 | Calcium-binding mitochondrial carrier protein Aralar2 | Other functions | 142 | 4 | 4 | 8,93 | 74102 |
| 4 | gi|61098372 | 60 kDa heat shock protein, mitocondrial | Chaperone; Cellular defense |
122 | 5,2 | 3 | 5,72 | 60972 |
| 5 | gi|118109616 | PREDICTED: similar to mitochondrial ATP synthase alpha subunit, partial | Oxidative phosphorylation | 77 | 29,2 | 2 | 5,8 | 5292 |
| 6 | gi|71897237 | ATP synthase subunit beta, mitochondrial precursor | Oxidative phosphorylation | 988 | 27,4 | 20 | 5,59 | 56627 |
| 7 | gi|50754375 | PREDICTED: cytochrome b-c1 complex subunit 1, mitochondrial | Oxidative phosphorylation | 145 | 4,6 | 4 | 6,58 | 52758 |
| 8 | gi|118098350 |
PREDICTED: cytochrome b-c1 complex subunit 2, mitochondrial-like isoform X4 |
Oxidative phosphorylation | 458 | 19 | 11 | 9,04 | 48579 |
| 10 | gi|71895153 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | Oxidative phosphorylation | 350 | 14,4 | 8 | 6,15 | 41431 |
| 11 | gi|57529307 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | Oxidative phosphorylation | 474 | 20,1 | 12 | 9,4 | 43079 |
| 13 | gi|46048903 | Voltage-dependent anion-selective channel protein 2 | Transport and carrier protein | 334 | 19,1 | 6 | 8,61 | 30197 |
| gi|124249322 | Prohibitin-2 | Regulates cell proliferation. May play a role in regulating mitochondrial respiration activity | 302 | 14 | 8 | 9,89 | 33336 | |
| 14 | gi|295148230 | Prohibitin | Regulates cell proliferation. May play a role in regulating mitochondrial respiration activity | 207 | 12,5 | 6 | 5,57 | 29892 |
| gi|196049778 | Chain D, Chicken Citocromo Bc1 complex Inhibited By An Iodinated Analogue Of The PolyketideCrocacin-d | Oxidative phosphorylation | 164 | 10,8 | 4 | 6,32 | 26939 | |
| gi|22775582 | ATP/ADP antiporter | Transport and carrier protein | 105 | 6,7 | 3 | 9,78 | 32847 | |
| gi|118089692 | PREDICTED: similar to ADP/ATP translocase | Transport and carrier protein | 105 | 7,6 | 3 | 9,72 | 29307 | |
| gi|54020693 | ADP/ATP translocase 3 | Transport and carrier protein | 102 | 6,7 | 3 | 9,73 | 32748 | |
| 15 | gi|226437575 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | Oxidative phosphorylation | 273 | 17,1 | 8 | 6,55 | 29232 |
| 16 | gi|118086790 | PREDICTED: similar to NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial precursor (NADH-ubiquinone oxidoreductase 24 kDa subunit) (NADH dehydrogenase subunit II) isoform 1 | Oxidative phosphorylation | 308 | 22,4 | 7 | 7,6 | 26893 |
| gi|5834847 | Cytochrome c oxidase subunit II (mitochondrion) | Oxidative phosphorylation | 127 | 9,3 | 3 | 4,57 | 25568 | |
| gi|50755667 | PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | Oxidative phosphorylation | 62 | 6,3 | 2 | 5,98 | 20497 | |
| 17 | gi|118103240 | PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like | Oxidative phosphorylation | 120 | 8,6 | 4 | 10,02 | 20473 |
| gi|118102465 | PREDICTED: ATP synthase subunit b, mitochondrial | Oxidative phosphorylation | 107 | 8,5 | 2 | 9,34 | 31751 | |
| lgi|118090950 | PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial | Oxidative phosphorylation | 104 | 11 | 2 | 5,84 | 23824 | |
| gi|118085386 | PREDICTED: chondroitin sulfate glucuronyltransferase-like | Tissue development and morphogenesis | 45 | 1,4 | 1 | 5,81 | 83804 | |
| 18 | gi|50745451 | PREDICTED: similar to ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d isoform 3 (BLAST: PREDICTED: ATP synthase subunit d, mitochondrial isoform 1 ) | Oxidative phosphorylation | 313 | 30,4 | 8 | 8,73 | 18343 |
| gi|118099484 | PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | Oxidative phosphorylation | 97 | 11,6 | 4 | 8,42 | 20125 | |
| 19 | gi|118099484 | PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | Oxidative phosphorylation | 91 | 11,6 | 4 | 8,42 | 20125 |
| gi|118083809 | PREDICTED: similar to LOC446923 protein isoform 1 (BLAST: ATP sinthasesubunidade O, isoformamitocondrial 2) | Oxidative phosphorylation | 66 | 6,7 | 2 | 9,88 | 22803 | |
| 20 | gi|296090732 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 | Oxidative phosphorylation | 165 | 27,4 | 6 | 9,57 | 16892 |
| gi|57529832 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial precursor | Oxidative phosphorylation | 151 | 18,3 | 6 | 7,9 | 21945 | |
Table 2: Summary of proteins from 2D BN / SDS Gels Identified by MALDI-TOF-TOF in chicken brain mitochondria.
| Pre-complex II | ||||||||
|---|---|---|---|---|---|---|---|---|
| Spot | Accession NCBI | Name of protein | Function | Mascot score | Sequence coverage (%) | Matches MS/MS | Theoretical | |
| pI | Mr | |||||||
| 1 | gi|57530041 | Mitochondrial inner membrane protein | Other functions | 586 | 12,7 | 13 | 5,72 | 79249 |
| gi|45383738 | aconitatehydratase, mitochondrial | Metabolic enzyme (TCA); mitochondrial DNA stability | 52 | 1,9 | 1 | 8,05 | 85790 | |
| 2 | gi|57529753 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | Oxidative phosphorylation | 766 | 16,1 | 16 | 6,5 | 79576 |
| 3 | gi|61098372 | 60 kDa heat shock protein, mitocondrial precursor | Chaperone; Cellular defense | 49 | 3,1 | 1 | 5,72 | 60972 |
| gi|45383566 | ATP synthase subunit alpha, mitochondrial | Oxidative phosphorylation | 40 | 2,4 | 2 | 9,29 | 60186 | |
| 4 | gi|118084029 | PREDICTED: uncharacterized protein LOC418583 | 34 | 0,9 | 1 | 5,64 | 146912 | |
| gi|118097244 | PREDICTED: dedicator of cytokinesis protein 2 | Modulates microglia secretion, phagocytosis and paracrine neurotoxicity | 34 | 0,7 | 1 | 8,3 | 219618 | |
| 5 | gi|71897237 | ATP synthase subunit beta, mitochondrial precursor | Oxidative phosphorylation | 887 | 23,5 | 17 | 5,59 | 56627 |
| 6 | gi|196049775 | Chain A, Chicken Cytochrome Bc1 complexo Inhibited By An Iodinated Analogue Of The PolyketideCrocacin-D | Oxidative phosphorylation | 406 | 12,6 | 10 | 5,95 | 49441 |
| 7 | gi|118098350 | PREDICTED: cytochrome b-c1 complex subunit 2, mitochondrial-like isoform X4 | Oxidative phosphorylation | 577 | 19 | 11 | 9,04 | 48579 |
| 8 | gi|45382953 | Aspartate aminotransferase, mitochondrial precursor | Amino acid metabolism | 633 | 19,4 | 12 | 9,38 | 47241 |
| 9 | gi|71895153 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | Oxidative phosphorylation | 455 | 18,6 | 9 | 6,15 | 41431 |
| 10 | gi|50758110 | PREDICTED: malate dehydrogenase, mitochondrial | Metabolic enzyme (TCA); | 318 | 12,5 | 5 | 8,83 | 36968 |
| gi|57529615 | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor | Ketogenesis pathway | 61 | 2,9 | 2 | 8,37 | 38237 | |
| gi|46048903 | voltage-dependent anion-selective channel protein 2 | Transport and carrier protein | 46 | 3,9 | 1 | 8,61 | 30197 | |
| 11 | gi|46048903 | voltage-dependent anion-selective channel protein 2 | Transport and carrier protein | 464 | 31,1 | 9 | 8,61 | 30197 |
| gi|124249322 | Prohibitin-2 | Regulates cell proliferation. May play a role in regulating mitochondrial respiration activity | 140 | 13,6 | 6 | 9,89 | 33336 | |
| 12 | gi|118123062 | PREDICTED: ATP synthase subunit gamma, mitochondrial isoform 1 | Oxidative phosphorylation | 269 | 11,3 | 6 | 9,39 | 32608 |
| gi|295148230 | Prohibitin | Regulates cell proliferation. May play a role in regulating mitochondrial respiration activity | 187 | 12,5 | 6 | 5,57 | 29892 | |
| 13 | gi|22775582 | ATP/ADP antiporter | Transport and carrier protein | 206 | 10,7 | 4 | 9,78 | 32847 |
| gi|54020693 | ADP/ATP translocase 3 | Transport and carrier protein | 204 | 10,7 | 4 | 9,73 | 32748 | |
| gi|57530120 | ADP/ATP translocase 1 | Transport and carrier protein | 97 | 8,4 | 2 | 9,74 | 32968 | |
| 14 | gi|226437575 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial | Oxidative phosphorylation | 561 | 27,2 | 14 | 6,55 | 29232 |
| 16 | gi|5834847 | cytochrome c oxidase subunit II (mitochondrion) | Oxidative phosphorylation | 287 | 25,1 | 8 | 4,57 | 25568 |
| gi|50755667 | PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | Oxidative phosphorylation | 155 | 7,4 | 4 | 5,98 | 20497 | |
| gi|118086790 | PREDICTED: similar to NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial precursor (NADH-ubiquinone oxidoreductase 24 kDa subunit) (NADH dehydrogenase subunit II) isoform 1 | Oxidative phosphorylation | 121 | 9,8 | 3 | 7,6 | 26893 | |
| 17 | gi|118103240 | PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like | Oxidative phosphorylation | 125 | 8,6 | 4 | 10,02 | 20473 |
| gi|118102465 | PREDICTED: ATP synthase subunit b, mitochondrial | Oxidative phosphorylation | 71 | 4,2 | 1 | 9,34 | 31751 | |
| gi|118090950 | PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial | Oxidative phosphorylation | 56 | 5,7 | 2 | 5,84 | 23824 | |
| 18 | gi|50745451 | PREDICTED: similar to ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d isoform 3 (BLAST: PREDICTED: ATP synthase subunit d, mitochondrial isoform 1 ) | Oxidative phosphorylation | 324 | 30,4 | 8 | 8,73 | 18343 |
| gi|118099484 | PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | Oxidative phosphorylation | 59 | 7,6 | 2 | 8,42 | 20125 | |
| gi|118083809 | PREDICTED: similar to LOC446923 protein isoform 1 (BLAST: ATP sinthasesubunidade O, isoformamitocondrial 2 ) | Oxidative phosphorylation | 46 | 6,7 | 2 | 9,88 | 22803 | |
| 19 | gi|118083809 | PREDICTED: similar to LOC446923 protein isoform 1 (BLAST: ATP sinthase subunit O, isoformamitocondrial 2 ) | Oxidative phosphorylation | 144 | 11,9 | 3 | 9,88 | 22803 |
| gi|118099484 | PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | Oxidative phosphorylation | 50 | 7,6 | 2 | 8,42 | 20125 | |
| 20 | gi|296090732 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 | Oxidative phosphorylation | 142 | 15,8 | 2 | 9,57 | 16892 |
| gi|57529832 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial precursor | Oxidative phosphorylation | 116 | 11 | 2 | 7,9 | 21945 | |
Table 3: Summary of proteins from 2D BN / SDS Gels Identified by MALDI-TOF-TOF in chicken brain mitochondria (Pre-complex II).
| Pre-complex III | ||||||||
|---|---|---|---|---|---|---|---|---|
| Spot | Accession NCBI | Name of protein | Function | Mascot score | Sequence coverage (%) | Matches MS/MS | Theoretical | |
| pI | Mr | |||||||
| 1 | gi|57530041 | Mitochondrial inner membrane protein | Other functions | 344 | 8,2 | 10 | 5,72 | 79249 |
| 2 | gi|57529753 | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | Oxidative phosphorylation | 90 | 4 | 4 | 6,5 | 79576 |
| gi|45384238 | Trifunctional enzyme subunit alpha, mitochondrial | Lipid metabolism; fatty acid beta-oxidation | 39 | 1,4 | 2 | 9,19 | 83186 | |
| 3 | gi|61098372 | 60 kDa heat shock protein, mitocondrial precursor | Chaperone; Cellular defense | 184 | 5,2 | 3 | 5,72 | 60972 |
| gi|45383566 | ATP synthase subunit alpha, mitochondrial | Oxidative phosphorylation | 35 | 2,4 | 1 | 9,29 | 60186 | |
| 4 | gi|45383566 | ATP synthase subunit alpha, mitochondrial | Oxidative phosphorylation | 658 | 17,5 | 12 | 9,29 | 60186 |
| 5 | gi|71897237 | ATP synthase subunit beta, mitochondrial precursor | Oxidative phosphorylation | 886 | 23,5 | 16 | 5,59 | 56627 |
| 6 | gi|50754375 | PREDICTED: cytochrome b-c1 complex subunit 1, mitochondrial | Oxidative phosphorylation | 273 | 9,2 | 9 | 6,58 | 52758 |
| gi|196049775 | Chain A, Chicken Cytochrome Bc1 complexo Inhibited By An Iodinated Analogue Of The PolyketideCrocacin-D | Oxidative phosphorylation | 273 | 9,9 | 9 | 5,95 | 49441 | |
| 7 | gi|118098350 | PREDICTED: cytochrome b-c1 complex subunit 2, mitochondrial-like isoform X4 | Oxidative phosphorylation | 360 | 15,1 | 9 | 9,04 | 48579 |
| gi|71897237 | ATP synthase subunit beta, mitochondrial precursor | Oxidative phosphorylation | 72 | 2,6 | 1 | 5,59 | 56627 | |
| 8 | gi|45382953 | Aspartate aminotransferase, mitochondrial precursor | Aminoacid metabolism | 337 | 15,4 | 11 | 9,38 | 47241 |
| 9 | gi|50758110 | PREDICTED: malate dehydrogenase, mitochondrial | Metabolic enzyme (TCA) | 233 | 12,8 | 6 | 8,83 | 36968 |
| gi|46048903 | voltage-dependent anion-selective channel protein 2 | Transport and carrier protein | 57 | 7,1 | 2 | 8,61 | 30197 | |
| gi|57529615 | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor | Ketogenesis pathway | 45 | 7,1 | 3 | 8,37 | 38237 | |
| 10 | gi|46048903 | voltage-dependent anion-selective channel protein 2 | Transport and carrier protein | 584 | 24 | 9 | 8,61 | 30197 |
| 11 | gi|118123062 | PREDICTED: ATP synthase subunit gamma, mitochondrial isoform 1 | Oxidative phosphorylation | 300 | 11,3 | 8 | 9,39 | 32608 |
| gi|50734923 | PREDICTED: carboxymethylenebutenolidase homolog isoform 3 | Prodrugbioactivation | 40 | 4,9 | 2 | 6,45 | 28170 | |
| gi|22775582 | ATP/ADP antiporter | Transport and carrier protein | 35 | 3 | 2 | 9,78 | 32847 | |
| gi|54020693 | ADP/ATP translocase 3 | Transport and carrier protein | 35 | 3 | 2 | 9,73 | 32748 | |
| gi|118089692 | PREDICTED: similar to ADP/ATP translocase | Transport and carrier protein | 35 | 3,4 | 2 | 9,72 | 29307 | |
| 12 | gi|54020693 | ADP/ATP translocase 3 | Transport and carrier protein | 309 | 17,8 | 7 | 9,73 | 32748 |
| 14 | gi|5834847 | cytochrome c oxidase subunit II (mitochondrion) | Oxidative phosphorylation | 345 | 25,1 | 9 | 4,57 | 25568 |
| gi|50755667 | PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | Oxidative phosphorylation | 142 | 7,4 | 4 | 5,98 | 20497 | |
| gi|118086790 | PREDICTED: similar to NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial precursor (NADH-ubiquinone oxidoreductase 24 kDa subunit) (NADH dehydrogenase subunit II) isoform 1 | Oxidative phosphorylation | 106 | 9,8 | 3 | 7,6 | 26893 | |
| gi|118093103 | PREDICTED: thioredoxin-dependent peroxide reductase, mitochondrial isoform X4 | Antioxidant protein | 72 | 4,9 | 2 | 8,4 | 30992 | |
| 15 | gi|118102465 | PREDICTED: ATP synthase subunit b, mitochondrial | Oxidative phosphorylation | 239 | 11,6 | 5 | 9,34 | 31751 |
| gi|118103240 | PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial-like | Oxidative phosphorylation | 103 | 8,6 | 4 | 10,02 | 20473 | |
| gi|118085922 | PREDICTED: similar to multidrug resistance protein 1a | Transport protein, limit access of drug to the central nervous system |
42 | 1,3 | 1 | 8,87 | 148466 | |
| gi|50745451 | PREDICTED: similar to ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d isoform 3 (BLAST: PREDICTED: ATP synthase subunit d, mitochondrial isoform 1 ) | Oxidative phosphorylation | 40 | 6,2 | 2 | 8,73 | 18343 | |
| 16 | gi|50745451 | PREDICTED: similar to ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d isoform 3 (BLAST: PREDICTED: ATP synthase subunit d, mitochondrial isoform 1 ) | Oxidative phosphorylation | 354 | 30,4 | 7 | 8,73 | 18343 |
| gi|118083809 | PREDICTED: similar to LOC446923 protein isoform 1 (BLAST: ATP sinthasesubunidade O, isoformamitocondrial 2) | Oxidative phosphorylation | 58 | 6,7 | 2 | 9,88 | 22803 | |
| gi|118099484 | PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | Oxidative phosphorylation | 50 | 7,6 | 2 | 8,42 | 20125 | |
| gi|50737115 | PREDICTED: DNA endonuclease RBBP8 | Cell cycle progression, DNA repair and transcriptional regulation | 42 | 1,1 | 1 | 6,14 | 103530 | |
| 17 | gi|118083809 | PREDICTED: similar to LOC446923 protein isoform 1 (BLAST: ATP sinthasesubunidade O, isoformamitocondrial 2 ) | Oxidative phosphorylation | 154 | 11,9 | 4 | 9,88 | 22803 |
| 18 | gi|118124369 | PREDICTED: ATP synthase subunit delta, mitochondrial-like, partial | Oxidative phosphorylation | 100 | 35 | 2 | 4,75 | 4159 |
| gi|71895513 | citocromo c oxidase subunidade 4 isoforma 1, mitocondrial | Oxidative phosphorylation | 47 | 7 | 2 | 8,91 | 19631 | |
| gi|118083465 | PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 isoform X1 | Oxidative phosphorylation | 37 | 6,2 | 2 | 9,46 | 18701 | |
| gi|2959450 | Desmin | Cell morphology | 36 | 2,9 | 1 | 5,3 | 51663 | |
Table 4: Summary of proteins from 2D BN / SDS Gels Identified by MALDI-TOF-TOF in chicken brain mitochondria (Pre-complex III).
| Pre-complex IV | ||||||||
|---|---|---|---|---|---|---|---|---|
| Spot | Accession NCBI | Name of protein | Function | Mascot score | Sequence average (%) | Matches MS/MS | Theoretical | |
| pI | Mr | |||||||
| 1 | gi|57530041 | Mitochondrial inner membrane protein | Other functions | 314 | 8,2 | 9 | 5,72 | 79249 |
| gi|45383738 | Aconitatehydratase, mitochondrial | Metabolic enzyme (TCA); mitochondrial DNA stability | 106 | 3,8 | 3 | 8,05 | 85790 | |
| gi|45383566 | ATP synthase subunit alpha, mitochondrial | Oxidative phosphorylation | 53 | 2,4 | 2 | 9,29 | 60186 | |
| 2 | gi|61098372 | 60 kDa heat shock protein, mitocondrial precursor | Chaperone; Cellular defense | 468 | 11,7 | 8 | 5,72 | 60972 |
| gi|45383566 | ATP synthase subunit alpha, mitochondrial | Oxidative phosphorylation | 107 | 2,4 | 2 | 9,29 | 60186 | |
| 4 | gi|71897237 | ATP synthase subunit beta, mitochondrial precursor | Oxidative phosphorylation | 816 | 23,5 | 16 | 5,59 | 56627 |
| 5 | gi|118088850 | PREDICTED: similar to QN1 orf | Cell division | 53 | 0,6 | 2 | 5,86 | 170342 |
| 6 | gi|118098350 | PREDICTED: cytochrome b-c1 complex subunit 2, mitochondrial-like isoform X4 | Oxidative phosphorylation | 541 | 19 | 11 | 9,04 | 48579 |
| 7 | gi|45382953 | Aspartate aminotransferase, mitochondrial precursor | Aminoacid metabolism | 364 | 15,4 | 12 | 9,38 | 47241 |
| gi|71897237 | ATP synthase subunit beta, mitochondrial precursor | Oxidative phosphorylation | 76 | 2,6 | 2 | 5,59 | 56627 | |
| 8 | gi|46048903 | Voltage-dependent anion-selective channel protein 2 | Transport and carrier protein | 603 | 30,4 | 8 | 8,61 | 30197 |
| 9 | gi|118123062 | PREDICTED: ATP synthase subunit gamma, mitochondrial isoform 1 | Oxidative phosphorylation | 296 | 11,3 | 8 | 9,39 | 32608 |
| gi|50734923 | PREDICTED: carboxymethylenebutenolidase homolog isoform 3 | Prodrugbioactivation | 40 | 4,9 | 2 | 6,45 | 28170 | |
| 10 | gi|22775582 | ATP/ADP antiporter | Transport and carrier protein | 51 | 3 | 2 | 9,78 | 32847 |
| gi|54020693 | adenine nucleotide translocator 3 | Transport and carrier protein | 51 | 3 | 2 | 9,73 | 32748 | |
| gi|118089692 | PREDICTED: similar to ADP/ATP translocase | Transport and carrier protein | 51 | 3,4 | 2 | 9,72 | 29307 | |
| 12 | gi|5834847 | cytochrome c oxidase subunit II (mitochondrion) | Oxidative phosphorylation | 260 | 16,3 | 8 | 4,57 | 25568 |
| gi|118093103 | PREDICTED: thioredoxin-dependent peroxide reductase, mitochondrial isoform X4 | Antioxidant protein | 64 | 4,9 | 2 | 8,4 | 30992 | |
| 13 | gi|118102465 | PREDICTED: similar to ATP synthase subunidade b | Oxidative phosphorylation | 428 | 15,8 | 12 | 9,34 | 31751 |
| gi|50745451 | PREDICTED: similar to ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d isoform 3 (BLAST: PREDICTED: ATP synthase subunit d, mitochondrial isoform 1 ) | Oxidative phosphorylation | 66 | 6,2 | 2 | 8,73 | 18343 | |
| gi|45384060 | Myosin-11 | Transport of vesicles | 40 | 0,9 | 1 | 5,5 | 228891 | |
| gi|118085922 | PREDICTED: similar to multidrug resistance protein 1a | Transport protein, limit access of drug to the central nervous system | 40 | 1,3 | 1 | 8,87 | 148466 | |
| 14 | gi|50745451 | PREDICTED: similar to ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d isoform 3 (BLAST: PREDICTED: ATP synthase subunit d, mitochondrial isoform 1 ) | Oxidative phosphorylation | 345 | 30,4 | 8 | 8,73 | 18343 |
| gi|118083809 | PREDICTED: similar to LOC446923 protein isoform 1 (BLAST: ATP sinthasesubunidade O, isoformamitocondrial 2 ) | Oxidative phosphorylation | 56 | 6,7 | 2 | 9,88 | 22803 | |
| gi|50737115 | DNA endonuclease RBBP8 | Other functions | 45 | 1,1 | 1 | 6,14 | 103530 | |
| 15 | gi|118083809 | PREDICTED: similar to LOC446923 protein isoform 1 (BLAST: ATP sinthasesubunidade O, isoformamitocondrial 2 ) | Oxidative phosphorylation | 53 | 6,7 | 2 | 9,88 | 22803 |
| 16 | gi|118124369 | PREDICTED: ATP synthase subunit delta, mitochondrial-like, partial | Oxidative phosphorylation | 101 | 35 | 2 | 4,75 | 4159 |
| gi|71895513 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | Oxidative phosphorylation | 37 | 7 | 2 | 8,91 | 19631 | |
Table 5: Summary of proteins from 2D BN / SDS Gels Identified by MALDI-TOF-TOF in chicken brain mitochondria (Pre-complex IV).
| Pre-complex V | ||||||||
|---|---|---|---|---|---|---|---|---|
| Spot | Accession NCBI | Name of protein | Function | Mascot score | Sequence coverage (%) | Matches MS/MS | Theoretical | |
| pI | Mr | |||||||
| 1 | gi|118082834 | PREDICTED: mitochondrial 10-formyltetrahydrofolate dehydrogenase (Alternative name: Aldehyde dehydrogenase family 1 member L2 | Detoxification, cell death | 39 | 1,4 | 2 | 6,5 | 101470 |
| 2 | gi|45384238 | Trifunctional enzyme subunit alpha, mitochondrial | Lipid metabolism; fatty acid beta-oxidation | 181 | 5,5 | 7 | 9,19 | 83186 |
| 3 | gi|61098372 | 60 kDa heat shock protein, mitocondrial precursor | Chaperone; Cellular defense | 539 | 16,4 | 11 | 5,72 | 60972 |
| gi|4521320 | unnamed protein product (BLAST: xin actin-binding repeat-containing protein 1) | Protects actin filaments from depolymerization | 39 | 1,6 | 1 | 7,3 | 87986 | |
| 4 | gi|118090114 | PREDICTED: similar to WW, C2 and coiled-coil domain containing 2 | Celular death | 32 | 0,8 | 1 | 5,07 | 132717 |
| 5 | gi|118098350 | PREDICTED: cytochrome b-c1 complex subunit 2, mitochondrial-like isoform X4 | Oxidative phosphorylation | 207 | 10,5 | 7 | 9,04 | 48579 |
| 6 | gi|45382953 | Aspartate aminotransferase, mitochondrial precursor | Aminoacid metabolism | 207 | 13,5 | 10 | 9,38 | 47241 |
| 7 | gi|50758110 | PREDICTED: malate dehydrogenase, mitochondrial | Metabolic enzyme (TCA) | 195 | 17,4 | 7 | 8,83 | 36968 |
| gi|46048903 | voltage-dependent anion-selective channel protein 2 | Transport and carrier protein | 116 | 11 | 3 | 8,61 | 30197 | |
| gi|57529615 | D-beta-hydroxybutyrate dehydrogenase, mitochondrial precursor | Ketogenesis pathway | 48 | 2,9 | 2 | 8,37 | 38237 | |
| 8 | gi|46048903 | voltage-dependent anion-selective channel protein 2 | Transport and carrier protein | 492 | 31,1 | 10 | 8,61 | 30197 |
| 9 | gi|22775582 | ATP/ADP antiporter | Transport and carrier protein | 251 | 13,4 | 7 | 9,78 | 32847 |
| gi|54020693 | adenine nucleotide translocator 3 | Transport and carrier protein | 249 | 13,4 | 7 | 9,73 | 32748 | |
| gi|57530120 | ADP/ATP translocase 1 | Transport and carrier protein | 185 | 11,1 | 5 | 9,74 | 32968 | |
| gi|57525378 | Phosphate carrier protein, mitochondrial | Transport and carrier protein | 58 | 3,6 | 2 | 9,33 | 37421 | |
| 10 | gi|5834847 | Cytochrome c oxidase subunit II (mitochondrion) | Oxidative phosphorylation | 248 | 16,3 | 8 | 4,57 | 25568 |
| gi|118093103 | PREDICTED: thioredoxin-dependent peroxide reductase, mitochondrial isoform X4 | Antioxidant protein | 148 | 8,7 | 4 | 8,4 | 30992 | |
| 11 | gi|71895513 | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | Oxidative phosphorylation | 469 | 36,3 | 14 | 8,91 | 19631 |
Table 6: Summary of proteins from 2D BN / SDS Gels Identified by MALDI-TOF-TOF in chicken brain mitochondria (Pre-complex V).
Twenty eight identified non-OXPHOS proteins were interacting with VDAC in avian brain mitochondria. The identified components of VDAC interacting chicken brain proteins in precomplex V (Table 1 and Figure 2) are ADP / ATP Translocase 1 (ANT1); Adenine Nucleotide Translocator 3 (ANT3); Mitochondrial 10-formyltetrahydrofolate dehydrogenase (AL1L2); ATP / ADP Antiporter (avANT); Aspartate Aminotransferase (AATM); D-Beta-Hydroxybutyrate Dehydrogenase (BDH1); 60 kDa heat shock protein (CH60); Cytochrome C Oxidase subunit II (COXII); Cytochrome C Oxidase subunit 4 Isoform 1 (COX4I1); Trifunctional enzyme subunit alpha (HADHA); Malate Dehydrogenase Mitochondrial (MDHM); unnamed protein product (BLAST: xin actin-binding repeat-containing protein 1, XIRP1); Voltage-Dependent Anion-selective Channel Protein 2 (VDAC2); Thioredoxin-dependent Peroxide Reductase Prdx3 protein (PRDX3); Phosphate Carrier Protein Mitochondrial (MPCP); Cytochrome b-c1 complex subunit 2, mitochondrial-like isoform X4 (QCR2) and similar to WW, C2 and coiled-coil domain containing 2 (WWC2). The proteins AL1L2, avANT, BDH1, CH60, COXII, COX4I1, HADHA, MDHM, PRDX3, XIRP1 and WWC2 were not VDAC interactants in bovine and rat brain mitochondria [18].
D-Beta-Hydroxybutyrate Dehydrogenase (BDH1) catalyzes the interconversion of acetoacetate and (R)-3-hydroxybutyrate, the two major ketone bodies produced during fatty acid catabolism. The authors showed that ketolytic (BDH1) and glycolytic enzymatic (Hexokinase) profiles of malignant brain tumors were different from the normal non-neoplastic brain tissue. A decrease in the mitochondrial enzymes BDH1 and OXCT1( 3-Oxoacid CoA Transferase) was coupled with positive expression of the glycolytic enzymes HK2 and PKM2 (Pyruvate Kinase M2 isoform), supporting the notion that many high grade brain tumors in humans have aberrant metabolism of ketones, and may preferentially use glucose for their energy needs [23].
PRDX3 is a mitochondrial antioxidant protein and a member of the peroxiredoxin family that can scavenge not only hydrogen peroxide (H2O2) in co-operation with thiol, but also peroxynitrite (ONOO–). PRDX3 markedly reduced gliosis, a post- neuronal cell death event and seems to be neuroprotective against oxidative insults [24].
Another VDAC interactant protein just observed in chicken brain mitochondria was xin actin-binding repeat-containing protein 1 (XIRP1). It was found as one of the proteins with an altered level in serum from schizophrenic patients [25]. The phosphate carrier protein (MPCP) was also found as a VDAC interactant in bovine brain mitochondria [18], and is a key component of the mitochondrial permeability transition pore (mPTP) [26], in the same way as VDAC [27]. MPCP undergoes a calcium-induced conformational change to induce pore formation.
In addition to the proteins identified in pre-complex V, fifteen other proteins were identified in pre-complex IV (Table 1 and Figure 2). They are Aconitate hydratase (ACON); similar to ADP / ATP translocase (ANT); ATP synthase subunit alpha (ATPA); ATP synthase subunit beta (ATPB); similar to ATP synthase subunit b (ATP5F1); ATP synthase subunit gamma (ATPG); ATP synthase subunit delta (ATPD); similar to ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d isoform 3 (ATP5H); similar to LOC446923 protein isoform 1 (BLAST: ATP synthase subunit O, mitochondrial isoform 2) (ATP5O); Carboxymethylenebutenolidase homolog isoform 3 (CMBL); DNA endonuclease RBBP8 (RBBP8); Myosin-11 (MYH11); Mitochondrial inner membrane protein (IMMT); similar to Multidrug Resistance Protein 1a (MRP1) and similar to QN1 orf (QN1). CMBL, RBBP8, MYH11, MRP1 and QN1 do not interact with VDACs in bovine and rat brain mitochondria [18]. Schizophrenic patients showed differences in activities of some enzymes from TCA cycle, like Aconitase (ACON) which presented a decreased activity [28]. The proteins ACON, F1-ATPase chains α (ATPA) and β (ATPB) were found differentially decreased in response to an acute hypobaric hypoxic episode and the subsequent re-oxygenation in rat brain cortex [29]. The authors suggest that these results could be due to the loss of proteins coupled with the destabilization of the mitochondria found after a hypobaric hypoxic insult, which would alter both the structure and functionality of ATPase and more specifically its catalytic subunit F1.
CMBL serves as a key enzyme in the activation of olmesartan medoxomil a prodrug type angiotensin II type I receptor antagonist. It is distributed in 20 tissues, including whole brain, but the highest level is found in liver [30]. And the protein RBBP8 or CtIP (C-terminal binding protein Interacting Protein) is a multifunctional protein involved in transcription, DNA replication, DNA repair by homologous recombination and the G1 and the G2 checkpoints. Both its functions and interactions point to a putative oncogenic potential of RBBP8 loss [31].
Our results demonstrated in avian brain several VDAC interactant proteins associated with neurological diseases, like the Multidrug resistance-associated protein 1 (MRP1). Drug resistance is one of the most serious problems in treatment of epilepsy [32]. Accumulating experimental evidence suggests that increased expression of MRP1 has been determined in epileptogenic brain regions of patients with pharmacoresistant epilepsy [33]. Over expression of such transporters in epileptogenic tissue is thus likely to reduce the amount of drug that reaches the epileptic neurons, which would be a likely explanation for pharmacoresistance [32].
In addition to the proteins identified in pre-complex IV, nine other proteins were identified in pre-complex III (Table 1 and Figure 2). They are Chain A, Chicken Cytochrome Bc1 complex Inhibited By An Iodinated Analogue Of The Polyketide Crocacin-D (3 CWB_A); Cytochrome b-c1 complex subunit 1 (QCRC1); Desmin, partial (DESM); NADH dehydrogenase [ubiquinone] 1 alpha sub complex subunit 8 (NDUFA8); NADH dehydrogenase [ubiquinone] 1 beta sub complex subunit 4 (NDUFB4); NADH dehydrogenase [ubiquinone] 1 beta sub complex subunit 10 (NDUFB10); NADH dehydrogenase [ubiquinone] iron-sulfur protein 7 (NDUFS7);
NADH-ubiquinone oxidoreductase 75 kDa subunit (NDUS1) and similar to NADH dehydrogenase [ubiquinone] flavoprotein 2 (NDUV2). Between these 8 proteins, just DESM was not interacting with VDACs in rat and bovine brain mitochondria [18]. Regarding molecular evidence of abnormal mitochondrial function in psychiatric disorders, in studies of schizophrenic patients, the 24 kDa and 51 kDa subunits of complex I from the electron transport chain were significantly decreased in the pre frontal cortex [34]. Mitochondrial dysfunction and abnormal brain bioenergetics have also been implicated in autism, with reduced expression of eleven genes of electron transport chain complex I [35].
Desmin plays an essential role in maintaining cell cytoarchitecture, positioning and functioning of organelles, and the intercellular signaling pathway [36]. Desmin was shown to regulate mitochondria affinity to ADP and oxygen consumption supposedly through direct binding to VDAC [37].
The identified components of pre-complex II, not found in precomplex III (Table 2) are uncharacterized protein LOC418583 (E1C825); Dedicator of cytokinesis protein 2 (DOCK2); NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10 (NDUAA); NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 (NDUFA12); NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8 (NDUFB8); NADH dehydrogenase [ubiquinone] iron-sulfur protein 8 (NDUFS8); NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 (NDUFS3); Prohibitin (PHB) and Prohibitin-2 (PHB2).
DOCK2 and E1C825 interact with VDACs just in chicken brain mitochondria [18]. DOCK2 is a guanyl nucleotide exchange factor expressed exclusively in brain microglia, and is regulated by PGE2 receptor EP2. It has been described that ablation of DOCK2 reduced amyloid burden in a model of Alzheimer´s disease [38].
PHBs have been functionally linked to diverse processes, such as transcriptional regulation, cell proliferation, development, and mitochondrial function [39]. Prohibitins exert a neuroprotective effect, by suppressing Reactive Oxygen Species (ROS) production and have a critical role in the activity of respiratory chain complex I [40]. It has been proposed that prohibitins promote longevity by moderating fat metabolism, mitochondrial proliferation and energy levels [41]. Its interaction with chicken brain VDAC, a gatekeeper in mitochondria mediated apoptosis is noteworthy.
In addition to proteins found in pre-complex II, four other proteins were identified in complex I. They are Calcium-binding mitochondrial carrier protein Aralar2 (Aralar2); Chondroitin sulfate glucuronyltransferase-like (CHPF2); Chain D Chicken Cytochrome Bc1 Complex Inhibited By An Iodinated Analogue Of The Polyketide Crocacin-D (3CWB_D) and NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9 (NDUA9).
Proteins Aralar2, CHPF2 and 3CWB_D interact with VDAC just in chicken mitochondria [18]. Aralar2 or Citrin is an aspartategluatamate carrier [42]. The presence of citrin in this study highlights a differential VDAC interactome in avian neuronal mitochondria in comparison with bovine and rat brains.
The protein CHPF2 is involved in glycosaminoglycan biosynthesis and plays a key role in tissue development and morphogenesis, and also contributes to tumor formation and development [43].
Analysis of the data reported above indicate that the kinetic assembly of protein complexes containing the VDAC follows a pattern quite different between chicken, bovine and rat brain [18]. The presence of ACON, Aralar2, CHPF2, CMBL, DOCK2, MRP1, PRDX3, RBBP8, Xirp1 and other proteins associated with VDAC only in chicken brain mitochondria, is in fact remarkable, and differentiate them from those of mammals, certainly in terms of developmental mechanisms of diseases, cell death and bioenergetics. Further studies are required to investigate if the differences in VDAC interactome reflect in differential metabolic and pathologic mechanisms between these species.
Acknowledgement
The work was supported by a grant from Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (2010/05560-6). Carla Rossini Crepaldi had a fellowship from Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (2009/06697-2). Biological samples were provided by Globoaves.
References
- BeltrandelRio H,WilsonJE (1991) Hexokinase of Rat Brain Mitochondria: Relative Importance of Adenylate Kinase and Oxidative Phosphorylation as Sources of Substrate ATP, and Interaction with Intramitochondrial Compartments of ATP and ADP. Arch BiochemBiophys 286: 183-194.
- Cerqueira César M, Wilson JE (2002) Functional Characteristics of Hexokinase Bound to the Type A and Type B Sites of Bovine Brain Mitochondria. Arch BiochemBiophys 397: 106-112.
- Kerner J, Lee K, Hoppel CL (2011) Post-translational modifications of mitochondrial outer membrane proteins. Free Radic Res 45: 16-28.
- Poleti MD, Tesch AC, Crepaldi CR, Souza GHMF, Eberlin MN, et al. (2010) Relationship Between Expression of Voltage-Dependent Anion Channel (VDAC) Isoforms and Type of Hexokinase Binding Sites on Brain Mitochondria. J MolNeurosci 41: 48-54.
- Shoshan-Barmatz V, Ben-Hail D (2012) VDAC, a multi-functional mitochondrial protein as a pharmacological target. Mitochondrion 12: 24-34.
- Chen C, Ko Y,Delannoy M,Ludtke S J, Chiu W, et al. (2004) Mitochondrial ATP synthasome: three-dimensional structure by electron microscopy of the ATP synthase in complex formation with carriers for Pi and ADP/ATP, J BiolChem 279: 31761-31768.
- Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, et al. (2010)VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med 31: 227-285.
- Abu-Hamad S, Sivan S, Shoshan-Barmatz V (2006)The expression level of the voltage-dependent anion channel controls life and death of the cell Proc. NatlAcadSci USA103: 5787-5792.
- Jiang W, Du B, Chi Z, Ma L, Wang S,et al. (2007) Preliminary explorations of the role of mitochondrial proteins in refractory epilepsy: some findings from comparative proteomics. J Neurosci Res 85: 3160-3170.
- Verna N, Pink M,Petrat F, Rettenmeyer AW, Scmitz-Spanke S (2015) Proeomic analysis of human Bladder epithelial cells by 2D Blue Native SDS-PAGE reveals TCDD-induced alterations of calcium and iron homeostasis possibly mediated by nitric oxide. J Proteome Res14: 202-213.
- Li F, Liang J, Wang W, Zhou X, Deng Z, et al. (2014) Analysis of Streptomyces coelicolor membrane proteome using two-dimensional native/native and native/sodium dodecyl sulfate gel electrophoresis. Anal Biochem 465: 148-155.
- Lasserre JP, Sylvius L, Joubert-Caron R, Caron M, Hardouin J (2010) Organellarprotein complexes ofcaco-2 human cells analyzed by two-dimensional blue native SDS-PAGE and mass spectrometry. J Proteome Res 9: 5093-5107.
- Peng Y, Luo Y, Yu T, Xu X, Fan K, et al.(2011) A Blue Native-PAGE analysis of membrane protein complexes in Clostridium thermocellum. BMC Microbiol 11: 22-32.
- Wöhlbrand L, Ruppersberg HS,Feenders C, Blasius B, Braun HP, et al. (2016)Analysis of membrane-protein complexes of the marine sulfate reducer Desulfobacula toluolica Tol2 by 1D blue native-PAGE complexome profiling and 2D blue native-/SDS-PAGE. Proteomics16: 973-988.
- Glew MD, Veith PD, Chen D, Seers CA, Chen YY, et al. (2014) Blue-native-PAGE analysis of membrane protein complexes in Porphyromonasgingivalis.J of Proteomics 110: 72-92.
- Van Gestel R A, van Solinge WW,vander Toorn HWP,Rijksen G, Heck AJR, et al. (2010) Quantitative erythrocyte membrane proteome analysis with blue-native/SDS PAGE. J Proteomics 73: 456-465.
- Basco D, Nicchia GP, Desaphy JF, Camerino DP, Frigeri A, et al. (2010) Analysis by two-dimensional blue native SDS-PAGE of membrane protein alterations in rat soleus muscle after hindlimb unloading. Eur J Appl Physiol 110, 1215-1224.
- Crepaldi CR, Vitale PAM, Tesch AC, Laure HJ, Rosa JC, et al. (2013) Application of 2D BN/SDS-PAGE coupled with mass spectrometry for identification of VDAC-associated protein complexes related to mitochondrial binding sites for type I brain hexokinase. Mitochondrion 13: 823-830.
- Schägger H, von Jagow G (1991) Blue Native Electrophoresis for Isolation of Membrane Protein Complexes in Enzymatically,Active Form, Anal. Biochem 199: 223-231.
- Schägger H,von JagowG (1987) Tricine-Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis for the Separation of Proteins in the Range from 1 kDato 100 kDa, Anal. Biochem 166: 368-379.
- Towbin H,Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. ProcNatlAcadSci USA 76: 4350-4354.
- Reisinger V, Eichaker LA (2007) How to analyze protein complexes by 2D Blue Native SDS-PAGE. Proteomics 7: 6-16.
- Chang HT, Olson LK, Schwartz KA (2013)Ketolytic and glycolytic enzymatic expression profiles in malignant gliomas: implication for ketogenic diet therapy. NutrMetab 10: 47.
- Hattori F, Murayama N,Noshita T,Oikawa S (2003) Mitochondrial peroxiredoxin-3 protects hippocampal neurons from excitotoxic injury in vivo. J Neurochem86: 860-868.
- Jaros JA, Martins-de-Souza D, Rahmoune H,Rothermundt M,Leweke FM, et al. (2012) Protein phosphorylation patterns in serum from schizophrenia patients and healthy controls. J Proteomics 76: 43-55.
- Varanyuwatana P, Halestrap A P (2012) The roles of phosphate and the phosphate carrier in the mitochondrial permeability transition pore. Mitochondrion 12: 120-125.
- Gincel D, Zaid H, Shoshan-Barmatz V (2001) Calcium binding and translocation by the voltage-dependent anion channel: a possible regulatory mechanism in mitochondrial function, Biochem. J 358: 147-155.
- Bubber P,Hartounian V, Gibson GE,Blass JP (2011) Abnormalities in the tricarboxylic acid (TCA) cycle in the brains of schizophrenia patients. EurNeuropsychopharmacol 21: 254-260.
- Hernández R,Blanco S, Peragón J,Pedrosa JA, Peinado MA (2013) Hypobaric hypoxia and reoxygenation induce proteomic profile changes in the rat brain cortex. Neuromolecular Med 15: 82-94.
- Ishizuka T, Fujimori I,Kato M,Noji-Sakikawa C, Saito M,et al. (2010) Human carboxymethylenebutenolidase as a bioactivating hydrolase of olmesartanmedoxomil in liver and intestine. J BiolChem 285: 11892-11902.
- Soria-Bretones I, Sáez C,Ruiz-Borrego M, Japón MA, Huertas P (2013) Prognostic value of CtIP/RBBP8 expression in breast câncer. Cancer Med 2: 774-783.
- Loscher W, Potschka H (2002)Role of Multidrug Transporters in Pharmacoresistance to Antiepileptic Drugs. J PharmacolExpTher 301: 7-14.
- Luna-TortósC, Fedrowitz M, Löscher W (2010) Evaluation of transport of common antiepileptic drugs by human multidrug resistance-associated proteins (MRP1, 2 and 5) that is overexpressed in pharmacoresistant epilepsy. Neuropharmacology 58 : 1019-1032.
- Clay HB, Sillivan S, Konradi C (2011) Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neuroscience 29: 311-324.
- Anitha A, Nakamura K, Thanseem I, Matsuzaki H, Miyachi T, et al. (2013) Downregulation of the expression of mitochondrial electron transport complex genes in autism. Brain Pathol 23: 294-302.
- Pawlak A, Gil RJ,GrajkowskaW,Nasierowska-Guttmejer AM, Rzezak J, et al. (2013) Significance of low desmin expression in cardiomyocytes in patients with idiopathic dilated cardiomyopathy. Am J Cardiol 111: 393-399.
- Rostovtseva TK, Bezrukov SM (2012) VDAC inhibition by tubulin and its physiological implications. BiochimBiophysActa 1818: 1526-1535.
- Cimino PJ,Yang Y,Li X, Hemingway JF, Cherne MK, et al. (2013)Ablation of the microglial protein DOCK2 reduces amyloid burden in a mouse model of Alzheimer's disease. ExpMolPathol 94: 366-371.
- Mishra S, Ande SR, Nyomba BLG (2010) The role of prohibitin in cell signaling. FEBS J 277: 3937-3946.
- Takahashi S, Masuda J, Shimagami H, Ohta Y,Kanda T, et al. (2011) Mild caloric restriction up-regulates the expression of prohibitin: A proteome study. Biochem and Biophys Res Comm 405: 462-467.
- Zhou P, Qian L, D'Aurelio M, Cho S, Wang G, et al. (2012)Prohibitin reduces mitochondrial free radical production and protects brain cells from different injury modalities. J Neurosci 32: 583-592.
- Contreras L,Urbieta A, Kobayashi K,Saheki T, Satrústegui J (2010)Low levels of citrin (SLC25A13) expression in adult mouse brain restricted to neuronal clusters. J Neurosci Res 88: 1009-1016.
- Kalathas D, Theocharis DA, Bounias D, Kyriakopoulou D,Papageorgakopoulou N, et al.(2011) Chondroitin synthases I, II, III and chondroitin sulfate lucuronyltransferase expression in colorectal cancer. Mol Med Rep 4: 363-368.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences